Key Points
We propose a novel oncogenic mechanism linked to the perinucleolar relocalization of chromosomal segments resulting from the translocation.
MCL and BL translocations result in new Ccnd1 and c-myc nuclear positioning, respectively, and nucleolin-dependent activation in both cases.
Abstract
In mantle cell lymphoma (MCL), one allele of the cyclin D1 (Ccnd1) gene is translocated from its normal localization on chromosome 11 to chromosome 14. This is considered as the crucial event in the transformation process of a normal naive B-cell; however, the actual molecular mechanism leading to Ccnd1 activation remains to be deciphered. Using a combination of three-dimensional and immuno-fluorescence in situ hybridization experiments, the radial position of the 2 Ccnd1 alleles was investigated in MCL-derived cell lines and malignant cells from affected patients. The translocated Ccnd1 allele was observed significantly more distant from the nuclear membrane than its nontranslocated counterpart, with a very high proportion of IgH-Ccnd1 chromosomal segments localized next to a nucleolus. These perinucleolar areas were found to contain active RNA polymerase II (PolII) clusters. Nucleoli are rich in nucleolin, a potent transcription factor that we found to bind sites within the Ccnd1 gene specifically in MCL cells and to activate Ccnd1 transcription. We propose that the Ccnd1 transcriptional activation in MCL cells relates to the repositioning of the rearranged IgH-Ccnd1–carrying chromosomal segment in a nuclear territory with abundant nucleolin and active PolII molecules. Similar transforming events could occur in Burkitt and other B-cell lymphomas.
Introduction
Many non-Hodgkin B-cell tumors are strongly associated with recurrent reciprocal chromosomal translocations. These include follicular lymphoma (FL), Burkitt’s lymphoma (BL), and mantle cell lymphoma (MCL), 3 malignancies in which the rearrangement most often implicates the immunoglobulin heavy chain (IgH) locus on chromosome 14 along with the Bcl2 antiapoptotic gene, the c-myc oncogene, or the cyclin D1 (Ccnd1) gene, respectively. Contrasting with the basal expression level of c-myc, both Bcl2 and Ccnd1 are silent in normal B cells. The molecular mechanisms at work in these various B-cell proliferations vary in relation with the stage of maturation of the concerned B lymphocyte as well as with the location of the cell when the initial cell transformation event occurs.1,2 Whereas BL and FL arise from germinal center B cells at various stages of maturation, MCL is believed to result from accidental IgH recombination events affecting B lymphocytes at the pre-B stage of differentiation in the bone marrow.3 Following the prototypical t(11;14)(q13;q32) rearrangement observed in the vast majority of MCL, one Ccnd1 gene allele becomes juxtaposed with the IgH locus on the derivative chromosome 14 (der14) and Ccnd1 gets constitutively expressed.4 It is generally assumed that this relies on the action of the potent Eµ enhancer present upstream of Ccnd1 in the IgH locus. The distance between the IgH and Ccnd1 genes can reach several hundreds of kilobases,5,6 however, and the question remains as to the actual molecular basis for Ccnd1 activation in MCL cells.
In a cell nucleus, intact chromosomes occupy specific territories.7,8 As a rule, but with a few exceptions,9 gene-dense chromosomes and actively transcribed chromosomal segments packaged in euchromatin tend to be more centrally located than gene-scarce, transcriptionally inactive heterochromatin segments.10-12 Chromosomal rearrangements such as those observed in hematopoietic malignancies necessarily trigger changes in the nuclear positioning of numerous genes.
Here, we have investigated the consequences of the chromosomal translocation on the position of the 2 alleles of the IgH and Ccnd1 genes in the nucleus of MCL cells. The IgH and Ccnd1 genes juxtaposed on the der14 were found more centrally located than the corresponding alleles on the nonrearranged chromosomes, and they were located very close to at least one nucleolus. Active RNA polymerase II (PolII) molecules were detected in these perinucleolar areas. We propose that the transcriptional activation of Ccnd1 in MCL cells directly relates to the nuclear repositioning of one Ccnd1 allele close to a nucleolus via Ccnd1 interaction with a nucleolus-abundant transcription factor nucleolin. Such a mechanism could also take place in other B-cell proliferations.
Materials and methods
Cells
The MCL-derived cell lines Granta-519, Jeko, UPN1, and UPN2 and the Raji and P3HR1 BL cell lines were grown as described in supplemental Methods. Patients’ and control lymphocytes were isolated from peripheral blood, separated by Ficoll-Hypaque density sedimentation, and placed in RPMI-1640 supplemented with 10% serum and antibiotics. HeLa and HeLaS3 cells were grown as described.13 Transient transfection of HeLa cells was performed using Lipofectamine2000 according to the manufacturer’s instructions.
Ethics statement
The human samples were obtained with the written consent of patients in accordance with the national legislation. This study was conducted in accordance with the Declaration of Helsinki.
3D-fluorescence in situ hybridization and immunodetection
Cells were immobilized on glass coverslips coated with Poly-d-lysine hydrobromide (Sigma). The cells were then treated as previously described to preserve their three-dimensional (3D) structure.14 Denatured nuclei were hybridized overnight with denatured probes, either IGH labeled green (RP11-346I20, AmpliTech) and CCND1 labeled orange (RP11-300I6, AmpliTech) for t(11;14) or IGH labeled green and MYC labeled red (RP11-80K22, AmpliTech) for t(8;14). After probe hybridization, slides were washed and nucleoli were detected using mouse anti-B23 antibody (Sigma) and goat anti-mouse Pacific Orange antibody (Invitrogen). Transcription factories or heterochromatin clusters were immunodetected simultaneously with detection of nucleoli using rabbit anti-RNA PolII phosphorylated at serine 5 (ActiveMotif) or rabbit anti-Histon H3 tri-methylated at lysine 9 (Upstate) antibodies, correspondingly, and goat anti-rabbit Alexa 633 (Invitrogen) antibody. DNA was counterstained with 4,6 diamidino-2-phenylindole (Vectashield, Vector) or Bobo1 (Invitrogen). Confocal microscopy, image processing, and statistical analysis were carried out as described elsewhere15-17 and in supplemental Methods.
Nuclear extracts and EMSA
Nuclear extracts were prepared from HeLaS3 cells, and electrophoretic mobility shift assay (EMSA) assay was carried out as described in supplemental Methods.
Plasmids
The pEGFPC1Nucleolin nucleolin expression plasmid expressing nucleolin18 was a kind gift of Dr J. Borowiec. The plasmids used for the luciferase activity were derived from the 1748D1Luc plasmid (a gift of Dr B. Sola), renamed pD1Luc. The putative LR1 sites were synthesized in vitro and cloned into the BamH1 restriction site of the 1748D1Luc: LR1.1 5′-GATCCTGGCCTCTCCCAGGCTGGGCCACCTGCCAGG-3′; LR1.2 5′-GATCCATCTCTCCCCAGCCTTGACCCCAATAAGG-3′; and LR1.3 5′-GATCCGAGCCATGGGTCCAGCCTGACCCGCATG-3′.
Reporter gene assays
Luciferase activity was determined 48 hours after transfection with luciferase reporter plasmid using the Dual Luciferase Assay (Promega) and normalized to protein concentration and to the activity of the phRL-TK reporter (Promega). All transfections were performed in triplicates and repeated in 3 independent experiments. Figures show the average result of 3 independent experiments.
Reverse transcription and quantitative polymerase chain reaction (PCR)
Total RNA was isolated from 2 × 106 HeLa cells transfected with the pEGFPC1Nucleolin expression plasmid18 and from mock-transfected HeLa cells using Trizol (Invitrogen) and reverse transcribed using the High Capacity cDNA Archive kit (Applied Biosystems). cDNA was amplified using specific primers and the Taqman PCR mix (Applied Biosystems). Expression was analyzed using the ΔΔCt method. The primers used for amplification are listed in supplemental Methods.
ChIP
Chromatin immunoprecipitation (ChIP) was performed starting from Granta-519 cells and normal human lymphocytes using the ChIP-IT Express kit (Active Motif) and antibodies against CTCF (Abcam) nucleolin (Sigma) and acetylated and methylated forms of H3 (Abcam) according to the manufacturer’s protocol. Immunoprecipitated DNA was PCR-amplified using specific primers listed in supplemental Methods.
ChIP-on-chip
DNA samples were hybridized to a human genome tiling array consisting of 50-mers positioned along nonrepeated sequences of the selected regions of chromosomes 8, 11, 14, 16, and X. Raw data were collected by Nimblegen (Roche-NimbleGen). The results were deposited in the GEO database. ChIP-on-chip analysis is described in supplemental Methods.
Results
The translocated Ccnd1 allele relocalizes close to a nucleolus
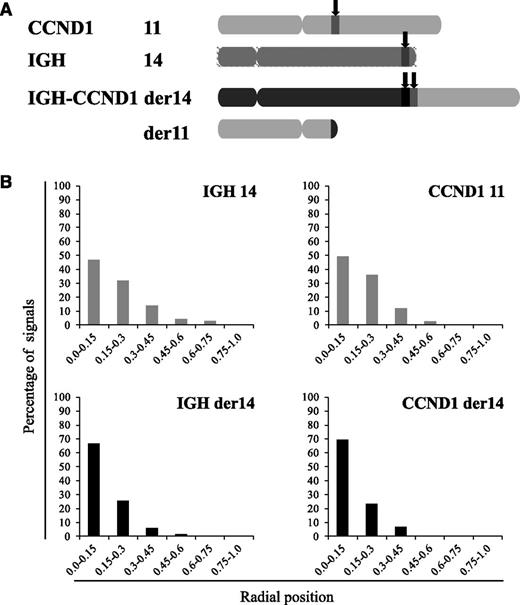
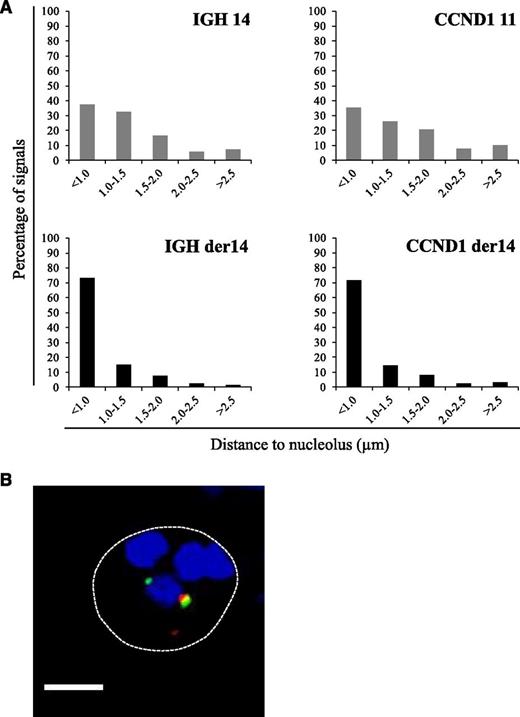
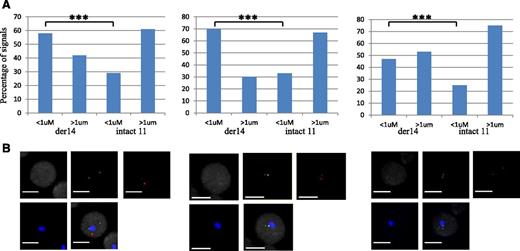
In MCL cells, the balanced t(11;14)(q14q32) translocation moves one Ccnd1 allele from its normal position on chromosome 11q to a position on the der14. We wondered whether this translocation was associated with a change in the nuclear positioning of the Ccnd1 and IgH genes. To analyze their radial positioning, cells from the MCL-derived Granta-519 cell line were analyzed by 3D-fluorescence in situ hybridization (FISH) using specific probes (Figure 1A). For each signal, the minimal distance to the nuclear membrane was measured in the 3-dimensional space of the nucleus. From the juxtaposed signals they produced, the IgH and Ccnd1 loci on the der14 were readily and systematically identified in every cell. Results reported in Figure 1B indicate that for each gene, the distribution of each of the 2 alleles differed according to whether Ccnd1 and IgH lay on a nonrearranged chromosome or on the der14. Thus, for both genes, the percentage of alleles in the centermost radial position was significantly higher when on the der14 than on the intact 14 (IgH, 67% vs 46%) or 11 (Ccnd1, 70% vs 49%) (P < .001). Similar results were obtained in UPN1 and Jeko, 2 other MCL cell lines tested, as well as in fresh lymphoma cells from MCL patients (supplemental Figure 1) as shown in Figure 3. Having observed this relocalization, we next asked whether the juxtaposed IgH and Ccnd1 alleles had been displaced toward a specific subnuclear domain. Indeed, the der14 IgH and Ccnd1 signals appeared to concentrate in the periphery of nucleoli, as previously noticed.19 To quantify this observation, distances to the closest nucleolus were measured for both alleles of the IgH and Ccnd1 genes. The distributions differed strikingly between normal and rearranged alleles (Figure 2; supplemental Figure 1). Thus, the der14 IgH and Ccnd1 signals were located <1 µm away from a nucleolus in >72% of the nuclei vs only ∼37% for their counterparts on intact chromosomes 11 and 14 (Figure 2; P < .001). Likewise, in lymphoma cells from 3 patients with MCL (Figure 3) as well as in the UPN1 and Jeko cell lines (supplemental Figure 1), the der14 Ccnd1 gene allele was found to lie close to the nucleolus much more frequently (58%, 70%, and 47% of the signals in each of the 3 patients) than the second allele on the nonrearranged chromosome (29%, 33%, and 24%, respectively; P < .001).
Repositioning of IgH and Ccnd1 in MCL cells. FISH was performed on 3-dimensionally preserved Granta-519 cell nuclei to reveal the IgH and Ccnd1 alleles. (A) Schematic representation of the chromosomes involved in the t(11;14) translocation in MCL cells and localization of the IgH and Ccnd1 probes used. (B) Distribution of the translocated (der14) and nontranslocated IgH and Ccnd1 alleles according to their radial positioning in Granta-519 cell nuclei. Distances were measured as described in Materials and methods. Histograms represent the percentage of FISH signals in the nuclear space with the range 0.0 to 0.15 corresponding to the centermost fraction of the nucleus and 0.75 to 1 to its periphery. Each graph represents a minimum of 120 FISH signals.
Repositioning of IgH and Ccnd1 in MCL cells. FISH was performed on 3-dimensionally preserved Granta-519 cell nuclei to reveal the IgH and Ccnd1 alleles. (A) Schematic representation of the chromosomes involved in the t(11;14) translocation in MCL cells and localization of the IgH and Ccnd1 probes used. (B) Distribution of the translocated (der14) and nontranslocated IgH and Ccnd1 alleles according to their radial positioning in Granta-519 cell nuclei. Distances were measured as described in Materials and methods. Histograms represent the percentage of FISH signals in the nuclear space with the range 0.0 to 0.15 corresponding to the centermost fraction of the nucleus and 0.75 to 1 to its periphery. Each graph represents a minimum of 120 FISH signals.
Perinucleolar relocalization of the der14 IgH and Ccnd1 alleles in the MCL Granta-519 cell line. The IgH and Ccnd1 alleles and nucleoli were revealed by immuno-FISH. (A) Distances to the closest nucleolus measured as described in Materials and methods for a minimum of 120 FISH signals. (B) A representative image of double-labeled DNA FISH in Granta-519 cell line. IgH is green, Ccnd1 is red, and nucleolus (B23) is blue. Scale bar represents 5 µm.
Perinucleolar relocalization of the der14 IgH and Ccnd1 alleles in the MCL Granta-519 cell line. The IgH and Ccnd1 alleles and nucleoli were revealed by immuno-FISH. (A) Distances to the closest nucleolus measured as described in Materials and methods for a minimum of 120 FISH signals. (B) A representative image of double-labeled DNA FISH in Granta-519 cell line. IgH is green, Ccnd1 is red, and nucleolus (B23) is blue. Scale bar represents 5 µm.
Perinucleolar relocalization of the translocated Ccnd1 allele in MCL patients. (A) Graphs show the distance of FISH signals to the nucleoli. Each graph corresponds to one patient with MCL. Distances have been measured in at least 100 nuclei. ***P < .001. (B) Representative images of double-labeled DNA FISH for each patient. IgH is green, Ccnd1 is red, and nucleolus (B23) is blue. Immuno-FISH and distance measurements were performed as in Figure 2, specifically on cells with the t(11;14) translocation, which were detected by colocalization of the IgH and Ccnd1 signals in the nuclei. Scale bar represents 5 μm.
Perinucleolar relocalization of the translocated Ccnd1 allele in MCL patients. (A) Graphs show the distance of FISH signals to the nucleoli. Each graph corresponds to one patient with MCL. Distances have been measured in at least 100 nuclei. ***P < .001. (B) Representative images of double-labeled DNA FISH for each patient. IgH is green, Ccnd1 is red, and nucleolus (B23) is blue. Immuno-FISH and distance measurements were performed as in Figure 2, specifically on cells with the t(11;14) translocation, which were detected by colocalization of the IgH and Ccnd1 signals in the nuclei. Scale bar represents 5 μm.
The rearranged IgH-Ccnd1–carrying chromosomal fragment associates with perinucleolar transcription factories, not heterochromatin
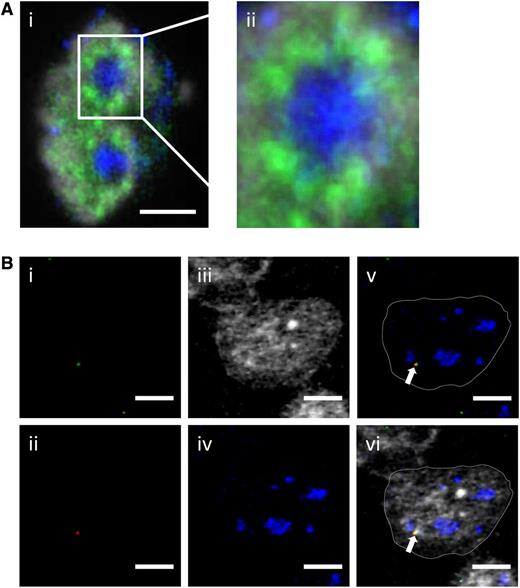
Ccnd1, which is not expressed in normal naive B cells, has been found expressed in a monoallelic fashion in cancer cells.19,20 Because the IgH-Ccnd1 chromosomal fragment on der14 chromosome is located close to a nucleolus, we wondered whether Ccnd1 transcription would actually take place in perinucleolar territories that are not known for being prone to PolII-dependent gene transcription. To test this, Granta-519 cells were stained using the B23 antibody that decorates nucleoli and an antibody to the phosphorylated C-terminal domain of the transcriptionally active form of PolII. Clusters of active PolII molecules were indeed readily detected in the immediate vicinity of nucleoli (Figure 4A). This was compatible with gene transcription taking place in this particular nuclear territory. In each cell, the proximity between the IgH-Ccnd1 chromosomal fragment and activated PolII foci was then evaluated. Signals from IgH (Figure 4Bi, green), Ccnd1 (Figure 4Bii, red), and PolII (Figure 4Biii, white) often coincided in a location immediately adjacent to a nucleolus (Figure 4Biv, blue). This can be clearly seen in the overlays superimposing the IgH, Ccnd1, and nucleolus with (Figure 4Bvi) or without (Figure 4Bv) the PolII staining. In contrast, the Ccnd1 signals generated from the intact chromosome 11 almost never colocalized with a PolII transcription factory (data not shown), consistent with the monoallelic expression of Ccnd1.
Simultaneous detection of fluorescently labeled transcription factories, nucleoli, heterochromatin, IgH, and Ccnd1 alleles in the Granta-519 MCL cell line. (A) (i) Active RNA PolII (green) and nucleoli (B23, blue) were immunostained with the nucleus counterstained with 4,6 diamidino-2-phenylindole (gray); (ii) enlargement focused on the nucleolus. Scale bar represents 3 μm. (B) IgH (green, i), Ccnd1 (red, ii), active RNA PolII (gray, iii), and nucleoli (B23, blue, iv) were simultaneously revealed by immuno-FISH. In merged images shown in (v) (merged from i, ii, and iv) and (vi) (merged from i, ii, iii, and iv), the arrow points to the der14-carried IgH and Ccnd1 alleles. Scale bar represents 3 µm.
Simultaneous detection of fluorescently labeled transcription factories, nucleoli, heterochromatin, IgH, and Ccnd1 alleles in the Granta-519 MCL cell line. (A) (i) Active RNA PolII (green) and nucleoli (B23, blue) were immunostained with the nucleus counterstained with 4,6 diamidino-2-phenylindole (gray); (ii) enlargement focused on the nucleolus. Scale bar represents 3 μm. (B) IgH (green, i), Ccnd1 (red, ii), active RNA PolII (gray, iii), and nucleoli (B23, blue, iv) were simultaneously revealed by immuno-FISH. In merged images shown in (v) (merged from i, ii, and iv) and (vi) (merged from i, ii, iii, and iv), the arrow points to the der14-carried IgH and Ccnd1 alleles. Scale bar represents 3 µm.
We then simultaneously visualized the IgH and Ccnd1 genes, nucleoli, and heterochromatin (H3K9me3). One example of such an analysis is displayed in supplemental Figure 3 with the 4 profiles generated along an axis shown in supplemental Figure 3i. As expected, the IgH (green) and Ccnd1 (red) peaks were almost superimposed, consistent with their juxtaposition on the der14. The nucleolus fluorescence (blue) lay very close to the IgH-Ccnd1 double peak. The heterochromatin fluorescence profile (gray) appeared as alternating peaks and ditches, with one ditch precisely corresponding to the IgH-Ccnd1 double peak. Taken together, these results confirmed the juxta-nucleolar localization of the IgH-Ccnd1 chromosomal fragment and its association with PolII transcription factories interspersed between heterochromatin-rich areas.
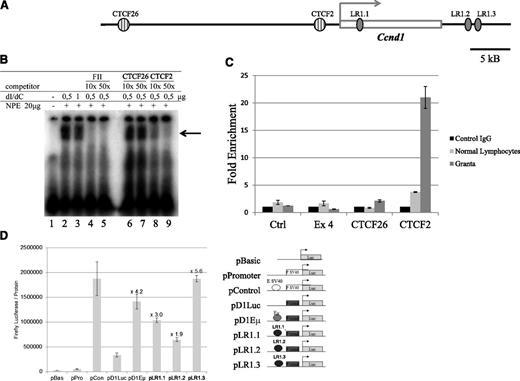
CTCF binds upstream of the Ccnd1 gene in MCL cells
Regardless of their subnuclear localization, gene loci can be protected from nearby activating elements by insulator sequences such as those bound by the CCCTC-binding factor CTCF (for review see Wallace and Felsenfeld21 ). Using the Transfac software,22 2 potential CTCF-binding sites, referred to here as CTCF26 and CTCF2, were identified 26 and 2 kb upstream of the Ccnd1 promoter (Figure 5A). Their potential as CTCF-binding sites was tested in EMSA experiments. A strong retardation signal was produced in the presence of CTCF2 (Figure 5B lanes 2-3). The electrophoretic retardation of labeled CTCF2 was abolished in the presence of a 10× excess of FII, the canonical CTCF site (lanes 4-5), or with cold CTCF2 (lanes 8-9) but not CTCF26 (lanes 6-7). The presence of CTCF was confirmed by ChIP experiments on Granta-519 cell line and normal lymphocytes. In control lymphocytes, the CTCF-specific antibody precipitated the chromatin fragment containing CTCF2, but not those containing CTCF26 or the negative control sequences (Figure 5C). This sharply contrasted with the ChIP results in Granta-519 cells, where a 21-fold enrichment was obtained with the CTCF2-containing chromatin fragment compared with <5-fold in normal lymphocytes. Thus, the CTCF-binding element located 2 kb upstream of the transcription start site of Ccnd1 is indeed occupied by CTCF in MCL cells and to a lesser extent in normal lymphocytes. This may play a role both in insulation of the Ccnd1 promoter and in transcriptional regulation via CTCF.
CTCF- and LR1-binding sites in the Ccnd1 gene. (A) Schematic representation of putative CTCF- and LR1-binding sites within and around the Ccnd1 gene. (B) Gel retardation assay using a labeled oligonucleotide corresponding to the CTCF2 sequence. dI/dC, unlabeled nonspecific competitor; FII, canonical CTCF site34 ; NPE, nuclear protein extract prepared from HeLa cell nuclei. CTCF2 and CTCF26 indicate unlabeled oligonucleotides tested for their capacity to compete for labeled CTCF2. The arrow points to the position of the CTCF2-specific band. (C) CTCF binding in vivo. ChIP was performed on Granta-519 cells and normal lymphocytes using anti-CTCF dark and light gray bars, CTCF (Ab) or a nonspecific antibody (black bars, ctrl IgG). The precipitated DNA fragments were analyzed by quantitative PCR using primers for amplification of sequences located ∼70 kb centromeric to the Ccnd1 gene or within its exon 4, neither being recognized by CTCF (immunoprecipitation negative controls, “ctrl” and “Ex4”); for CTCF26 and CTCF2, the 2 potential CTCF binding sites identified in Ccnd1. Error bars represent the standard deviation between duplicates within the same experiment. (D) LR1-binding sites in the Ccnd1 gene as transcriptional enhancers. The transcriptional activity of the 3 potential LR1-binding sites identified in the Ccnd1 gene was tested in HeLa cells 48 hours after transfection. The enhancer strength was quantified relative to the luciferase activity generated with the reference SV40 promoter.
CTCF- and LR1-binding sites in the Ccnd1 gene. (A) Schematic representation of putative CTCF- and LR1-binding sites within and around the Ccnd1 gene. (B) Gel retardation assay using a labeled oligonucleotide corresponding to the CTCF2 sequence. dI/dC, unlabeled nonspecific competitor; FII, canonical CTCF site34 ; NPE, nuclear protein extract prepared from HeLa cell nuclei. CTCF2 and CTCF26 indicate unlabeled oligonucleotides tested for their capacity to compete for labeled CTCF2. The arrow points to the position of the CTCF2-specific band. (C) CTCF binding in vivo. ChIP was performed on Granta-519 cells and normal lymphocytes using anti-CTCF dark and light gray bars, CTCF (Ab) or a nonspecific antibody (black bars, ctrl IgG). The precipitated DNA fragments were analyzed by quantitative PCR using primers for amplification of sequences located ∼70 kb centromeric to the Ccnd1 gene or within its exon 4, neither being recognized by CTCF (immunoprecipitation negative controls, “ctrl” and “Ex4”); for CTCF26 and CTCF2, the 2 potential CTCF binding sites identified in Ccnd1. Error bars represent the standard deviation between duplicates within the same experiment. (D) LR1-binding sites in the Ccnd1 gene as transcriptional enhancers. The transcriptional activity of the 3 potential LR1-binding sites identified in the Ccnd1 gene was tested in HeLa cells 48 hours after transfection. The enhancer strength was quantified relative to the luciferase activity generated with the reference SV40 promoter.
Nucleolin binds the Ccnd1 gene and activates its transcription
Because in MCL cells, the Ccnd1 gene is transcribed despite CTCF being bound immediately upstream of its transcription start site, we reasoned that a nucleolus-derived factor interacting with potential enhancer elements located further downstream in the Ccnd1 gene would be well suited to play a role in transcriptional activation. One of the most abundant components in nucleoli is nucleolin, a multifunctional protein implicated in transcription regulation and other nuclear processes. Together with HnRNP-D, nucleolin forms a site-specific DNA-binding heterodimer known as LR1, whose transactivating activity is crucial for normal B-cell development.23 During B-cell differentiation, intragenic recombination and class switch require the binding of LR1 to specific nucleotide sequences in the switch region of the IgH locus.24 We have detected 3 putative LR1-binding sites within or in the vicinity of the Ccnd1 gene, one intronic in the body of the gene (LR1.1), the other 2 (LR1.2 and LR1.3) immediately downstream of the Ccnd1 coding sequence (Figure 5A). Each site was cloned into a luciferase-expressing reporter and tested in HeLa cells for its capacity to enhance basal transcription. Luciferase expression increased 1.9- to 5.6-fold in cells transfected with each of the 3 LR1 site-containing constructs, comparable with the 4.2-fold increase obtained using the positive control Eµ enhancer from the IgH gene (Figure 5D). Thus, the nucleolin/LR1 binding sequences present in the Ccnd1 gene behave as bona fide transcriptional enhancers, consistent with the an activating role in Ccnd1 transcription in MCL cells.
To further investigate this possibility, ChIP-on-chip experiments were performed on Granta-519 MCL cells and normal lymphocytes using an anti-nucleolin antibody. An MCL-specific interaction was observed, the presence of nucleolin being testified at several locations within and around the Ccnd1 gene, including the above-described LR1 sites (supplemental Figure 2A-B). A higher nucleolin content was additionally observed near the breakpoint region on the rearranged chromosome 14, but not on a chromosome 8 region used as control (supplemental Figure 2C-D; supplemental Table 1). The ChIP-on-chip experiments also revealed vast amounts of lysine 9-acetylated histone H3 at the Ccnd1 promoter (supplemental Figure 2A-B), consistent with the high transcriptional activity of this gene. Together, these data are in agreement with the previously reported presence of active chromatin marks at the Ccnd1 promoter region in MCL cells4 and further indicate that the relocalization of the CCND1 locus favors its increased interaction with nucleolin.
A transactivating role for nucleolin in MCL and other non-Hodgkin B-cell lymphomas
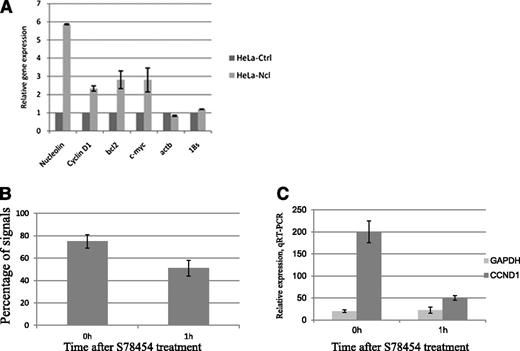
Thus, in MCL cells, the translocated Ccnd1 allele gets localized in a perinucleolar territory and becomes transcriptionally active, presumably under the influence of nucleolin. To further circumstantiate a transactivating role for this protein, HeLa cells were transfected with a green fluorescent protein (GFP)-nucleolin construct. GFP-positive cells were sorted and found to contain nucleolin spread throughout the nucleus (data not shown). In these transfectants, Ccnd1 transcription levels were more than 2-fold higher than those measured in mock-transfected cells, with actin B or rRNA 18S transcription remaining unchanged (Figure 6A). Of note, a nucleolin knockdown performed in MCL cells leads to massive apoptosis, precluding any relevant experiment.25 Thus, although highly likely, an essential role of nucleolin for Ccnd1 expression in MCL cells cannot be formally proven.
Ccnd1 transcription levels and colocalization of nucleoli and Ccnd1. (A) A nucleolin-GFP pEGFPC1. Nucelolin construct was transfected into HeLa cells followed by selection of GFP-positive cells by flow cytofluorimetry. Transcription of nucleolin, Ccnd1, c-myc, Bcl2, Actin B, and 18S rRNA studied by quantitative reverse-transcription PCR in GFP-nucleolin and mock-transfected cells. Error bars represent the standard deviation between duplicates. (B-C) Ccnd1 localization and expression in Granta-519 cells treated with a histone deacetylase inhibitor. The localization of a Ccnd1 allele <1 µm from a nucleolus (B) as determined using 3D-FISH and Ccnd1 transcription rate (C) as measured by quantitative reverse-transcription PCR. The data represent the average results from 2 independent experiments.
Ccnd1 transcription levels and colocalization of nucleoli and Ccnd1. (A) A nucleolin-GFP pEGFPC1. Nucelolin construct was transfected into HeLa cells followed by selection of GFP-positive cells by flow cytofluorimetry. Transcription of nucleolin, Ccnd1, c-myc, Bcl2, Actin B, and 18S rRNA studied by quantitative reverse-transcription PCR in GFP-nucleolin and mock-transfected cells. Error bars represent the standard deviation between duplicates. (B-C) Ccnd1 localization and expression in Granta-519 cells treated with a histone deacetylase inhibitor. The localization of a Ccnd1 allele <1 µm from a nucleolus (B) as determined using 3D-FISH and Ccnd1 transcription rate (C) as measured by quantitative reverse-transcription PCR. The data represent the average results from 2 independent experiments.
Whether the proximity with a nucleolus is important for Ccnd1 transcription in MCL cells was further tested when Granta-519 MCL cells were submitted to Abexinostat (S78454 / PCI-24781), a pan-histone deacetylase inhibitor whose action provokes an overall disorganization of the nuclear structure. In cells thus treated, the percentage of the der14-carried Ccnd1 alleles still localized next to a nucleolus decreased from 75% to 53% as early as 1 hour after treatment (Figure 6B), and this was accompanied by a 4-fold reduction in Ccnd1 transcription (Figure 6C), consistent with Ccnd1 expression in MCL cells requiring an unperturbed chromatin and nuclear organization.
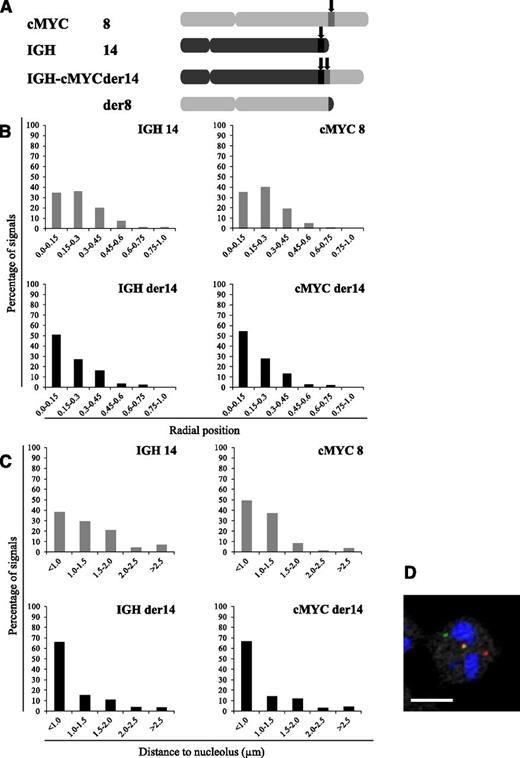
Bcl2 and c-myc, the 2 proto-oncogenes activated in follicular and BLs, respectively, were also found activated in nucleolin transfectants (Figure 6A). Confirming the previous report that the nucleolin-containing transcription factor LR1 is a transcriptional activator for c-myc,26 this further suggested that in these B-cell tumors with a t(14;18) or t(8;14) translocation, an important step in the transformation process could relate to the relocalization in the vicinity of a nucleolus of a fragment of the rearranged IgH-carrying chromosome. This hypothesis was tackled using the endemic BL-derived Raji and P3HR1 cell lines that harbor the canonical t(8;14)(q24;q32) translocation. In this cell line, one allele of the c-myc oncogene, normally on chromosome 8, gets localized immediately telomeric to an IgH switch region on the der14 following the translocation event. Our 3D-FISH results generated using specifically designed probes (Figure 7A; supplemental Figure 1) were very similar to those observed in MCL cells. Indeed, the IgH and c-myc alleles juxtaposed on the der14 were observed farther from the nuclear membrane than their counterparts on intact chromosomes 14 and 8 (34% vs 51% and 35% vs 54% centrally localized IgH and c-myc alleles, respectively; P < .01) (Figure 7B). Furthermore, the IgH and c-myc alleles on the rearranged chromosome 14 lay closer to a nucleolus than those on intact chromosomes 14 and 8 (IgH, 66% vs 38%, and c-myc, 67% vs 49% of the alleles on the rearranged vs nonrearranged chromosomes, respectively; P < .01) (Figure 7C).
Radial localization and distances to closest nucleolus for IgH and c-myc loci in the Raji BL cell line. (A) Schematic representation of the chromosomes involved in the t(8;14) translocation in the Raji BL cell line and localization of the IgH and c-myc probes used. (B) Distribution of the translocated (IgH – c-myc der14) and nontranslocated (IgH 14 and c-myc 8) IgH and c-myc alleles according to their radial positioning in Raji cell nuclei. Distances were measured as described in Materials and methods. Histograms represent the percentage of FISH signals in the nuclear space with the range 0.0 to 0.15 corresponding to the centermost fraction of the nucleus and 0.75 to 1 to its periphery. Each graph represents a minimum of 115 FISH signals. (C) Perinucleolar relocalization of the der14 IgH and c-myc alleles in the BL Raji cell line. The IgH and c-myc alleles and nucleoli were revealed by immuno-FISH. Distances to the closest nucleolus were measured as described in Materials and methods for a minimum of 115 FISH signals. (D) A representative image of double-labeled DNA FISH in RAJI cell line. IgH is green, c-myc is red, and nucleolus (B23) is blue. Scale bar represents 5 µm.
Radial localization and distances to closest nucleolus for IgH and c-myc loci in the Raji BL cell line. (A) Schematic representation of the chromosomes involved in the t(8;14) translocation in the Raji BL cell line and localization of the IgH and c-myc probes used. (B) Distribution of the translocated (IgH – c-myc der14) and nontranslocated (IgH 14 and c-myc 8) IgH and c-myc alleles according to their radial positioning in Raji cell nuclei. Distances were measured as described in Materials and methods. Histograms represent the percentage of FISH signals in the nuclear space with the range 0.0 to 0.15 corresponding to the centermost fraction of the nucleus and 0.75 to 1 to its periphery. Each graph represents a minimum of 115 FISH signals. (C) Perinucleolar relocalization of the der14 IgH and c-myc alleles in the BL Raji cell line. The IgH and c-myc alleles and nucleoli were revealed by immuno-FISH. Distances to the closest nucleolus were measured as described in Materials and methods for a minimum of 115 FISH signals. (D) A representative image of double-labeled DNA FISH in RAJI cell line. IgH is green, c-myc is red, and nucleolus (B23) is blue. Scale bar represents 5 µm.
Discussion
Experiments performed in the early and mid-1980s led to the idea that the translocation of a proto-oncogene next to a transcriptional activator would trigger molecular events crucial for cell transformation. Experiments with the c-myc proto-oncogene were particularly thought-provoking. Mice rendered transgenic with an unmodified c-myc placed immediately downstream of the IgH locus Eµ enhancer exhibited enlarged B-cell compartments resulting in the constant emergence of monoclonal B-cell proliferations.27 At the time, the constructs had been designed upon the naturally occurring rearrangements observed in mouse plasmacytomas. In those early transgenic experiments, the genetic elements introduced in murine recipients were separated by hundreds of nucleotides only, far different from the genomic organization observed in human translocation-bearing B-cell tumors where the distance between the IgH locus and the translocated proto-oncogene can be orders of magnitude larger. Such is indeed the case in the vast majority of FLs with the t(14;18) translocation. Following the chromosomal rearrangement, one allele of the Bcl2 antiapoptotic gene is displaced from chromosome 18 to the der14, where it now lays hundreds of kilobases telomeric to the IgH locus. Using the chromosome conformation capture methodology, Duan et al28 have evidenced a direct interaction between enhancers present in the 3′ region of the IgH locus and the 5′ promoter region of the newly transcribed translocated Bcl2 allele. This is in accordance with our improved understanding of chromatin loops and nuclear organization that allow regulatory elements positioned in cis at distances up to the megabase range to come in direct contact with each other in the 3-dimensional space of the cell nucleus (for review see Hou and Corces29 ).
Here, we have focused on MCL, a B-cell proliferation that has been less studied so far but whose chromosomal rearrangement also interests the IgH locus. Similar to Bcl2 in FL, the translocated Ccnd1 allele lies at a relatively large linear distance from transcriptional enhancers in the IgH locus. Also similarly, the translocation results in the expression of a gene that is silent in normal naive B cells and whose expression appears directly related to the transformation process. Importantly, MCL differs from the BL model, as transgenic mice expressing the Ccnd1 gene under the control of the IgH Eµ enhancer do not develop lymphomas.30 To better understand the underlying mechanisms induced by the t(11;14) translocation, we have explored the nuclear positioning of each allele of the Ccnd1 gene in MCL cells. In both cell lines and fresh tumor cells, the translocated Ccnd1 allele was observed farther from the nuclear membrane than the allele on the intact chromosome 11 and also farther than the usual Ccnd1 radial position in naive B cells (data not shown). This translocation-associated displacement away from the nuclear membrane resulted in a new positioning in the vicinity of a nucleolus, a difference that was statistically significant between the Ccnd1 alleles on the rearranged vs intact chromosomes. In MCL cells, Ccnd1 is known to be monoallelically expressed,20 a situation also reported for other somatic genes.31 Our data are totally consistent with transcription taking place solely from the translocated Ccnd1 allele.
Using immuno-FISH experiments, the IgH and Ccnd1 loci present on the der14 were revealed as juxtaposed signals. In most instances, those were located in the immediate vicinity of clusters of transcriptionally active PolII molecules, in sharp contrast with the untranslocated Ccnd1 alleles whose vast majority lay away from transcription factories. The presence of active PolII clusters in the periphery of nucleoli, which in normal cells is mostly occupied by heterochromatin and therefore not prone to PolII transcription, should not come as a surprise, because active chromatin marks have been reported previously in the Ccnd1 promoter region of MCL cells.4
What triggers Ccnd1 transcription from the der14? To answer this question, we hypothesized that a nucleolus-derived factor could contribute to this transcriptional activation. LR1 is a DNA-binding heterodimer composed of HnRNP-D and nucleolin, a major component in nucleoli and a potent transcriptional activator. Binding sites for LR1 were identified within the Ccnd1 gene and indeed shown to function as transcriptional enhancers. In ChIP experiments, nucleolin was directly detected in vivo within and around the Ccnd1 gene. Furthermore, Ccnd1 transcription was also activated in transfected HeLa cells overexpressing nucleolin. Conversely, when the overall organization of MCL Granta-519 cells was perturbed following treatment with an inhibitor of histone deacetylases, the percentage of Ccnd1 alleles associated with a nucleolus rapidly decreased in parallel with levels of Ccnd1 transcription. Together, these results strongly suggest that Ccnd1 transcription gets activated in MCL cells, because the translocated Ccnd1 allele gets localized in the nucleolin-abundant environment found in perinucleolar areas. In this regard, it is tempting to speculate that nucleolin and possibly other nucleolar components such as nucleophosmin or ribosomal proteins could also participate in PolII transcription in malignant cells. Interestingly, nucleophosmin associates with another factor, CTCF.32 We have observed MCL-specific binding of CTCF in the vicinity of the Ccnd1 promoter. Similar binding has been previously reported in BL cells.33 This binding may play a role both in the insulation of the Ccnd1 promoter and in its transcriptional regulation via CTCF.
The literature contains one report of inhibition of nucleolin expression in HeLa cells and fibroblasts treated for 5 days with a combination of small interfering RNA. This inhibition was accompanied by many abnormalities, including a lowered proliferation index, an accumulation of cells in the G2 phase of the cell cycle, and eventual entry of the cells into apoptosis.25 In addition to its ribosomal-related functions, nucleolin has also been characterized as a histone chaperone and chromatin co-remodeler. In view of its variable cellular localization and pleiotropic functions and because of the well-known difficulties for efficiently transfecting B lymphocytes, we have not attempted here to extinguish nucleolin expression in MCL cells.
The results we have obtained are partially contradictory to those reported by the Epner’s group,19 who found no difference in positioning between the translocated and untranslocated Ccnd1 alleles. Possible reasons for this discrepancy include our methodology to measure radial positioning and distances to a nucleolus. Here, we have carefully distinguished between the translocated and nontranslocated alleles, the latter being almost never observed in the proximity of a nucleolus. Also, the 3D-FISH methodology we have applied to MCL cell lines and malignant cells from MCL patients may have allowed more precise measurements than in the 2-dimensional FISH experiments performed by Liu et al.19
To account for Ccnd1 expression in MCL cells, we propose a model whereby the key event occurs when one Ccnd1 allele gets newly localized to a transcriptionally favorable perinucleolar territory in these cancer cells. Can a similar mechanism apply in other hematopoietic malignancies with chromosomal rearrangements interesting acrocentric chromosomes? In the major reciprocal t(8;14) translocation associated with Burkitt lymphomas, the chromosome 8-carried proto-oncogene c-myc gets localized head to head next to the IgH locus on the rearranged acrocentric 14. Differing from sporadic forms of BL where the 5′ end of c-myc is decapitated following the chromosomal break, in the endemic form, an intact c-myc gene gets juxtaposed to the IgH locus along with hundreds of kilobases of genomic sequence from the 5′ region of c-myc. In this new setting, c-myc is localized at a long distance from the IgH locus, reminiscent of the situation in FL and MCL. In Raji, a prototypical endemic BL cell line with c-myc hundreds of kilobases telomeric to the IgH locus, our experiments have revealed that the rearranged chromosome 14 fragment, which now carries IgH, and c-myc gets repositioned close to a nucleolus. In this new setting, c-myc may fall in the action range of nucleolar factors such as nucleolin, which in our transcription assays was found to activate c-myc even better than Ccnd1. Further experiments will be necessary to explore the actual relevance of such a mechanism in various B-cell proliferations and to investigate the respective role of nucleolar components and cis regulation in the activation of genes crucial for transformation in MCL, BL, and other non-Hodgkin B-cell lymphomas.
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr J. Borwiec and B. Sola for the gift of plasmids and the Laboratoires Servier for the gift of Abexinostat.
This research was supported by grants from the Fondation de France and Institut National de Cancer to Y.S.V., from Presidium of the Russian Academy of Sciences (MCB grant), and from Russian Foundation for Basic Research 12-04-93109 to S.R. and 13-04-93105 to O.I. A.P. was supported by a postdoctoral fellowship from the Fondation de France.
Authorship
Contribution: J.A., A.P., O.I., M.K., E.Z.-Z., D.M., and N.P. performed research; A.B., J.W., M.L., V.R., O.I., S.V.R., M.L., and Y.S.V. analyzed data; V.C.-C. and V.R. contributed reagents; and M.L. and Y.S.V. wrote the paper
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Yegor S. Vassetzky, UMR8126, Universite Paris-Sud, CNRS, Institut de Cancerologie Gustave Roussy, 39 Rue Camille Desmoulins, Villejuif Cedex, 94805 France; e-mail: vassetzky@igr.fr.
References
Author notes
J.A., A.P., and O.I. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal