Key Points
Carrying the KIR A/A genotype contributes to risk of childhood ALL, particularly in Hispanics.
Abstract
Killer cell immunoglobulin-like receptors (KIRs), via interaction with their cognate HLA class I ligands, play a crucial role in the development and activity of natural killer cells. Following recent reports of KIR gene associations in childhood acute lymphoblastic leukemia (ALL), we present a more in-depth investigation of KIR genes and their cognate HLA ligands on childhood ALL risk. Genotyping of 16 KIR genes, along with HLA class I groups C1/C2 and Bw4 supertype ligands, was carried out in 212 childhood ALL cases and 231 healthy controls. Frequencies of KIR genes, KIR haplotypes, and combinations of KIR-HLA ligands were tested for disease association using logistic regression analyses. KIR A/A genotype frequency was significantly increased in cases (33.5%) compared with controls (24.2%) (odds ratio [OR] = 1.57; 95% confidence interval [CI], 1.04-2.39). Stratifying analysis by ethnicity, a significant difference in KIR genotype frequency was demonstrated in Hispanic cases (34.2%) compared with controls (21.9%) (OR = 1.86; 95% CI, 1.05-3.31). Homozygosity for the HLA-Bw4 allele was strongly associated with increased ALL risk exclusively in non-Hispanic white children (OR = 3.93; 95% CI, 1.44-12.64). Our findings suggest a role for KIR genes and their HLA ligands in childhood ALL etiology that may vary among ethnic groups.
Introduction
Childhood acute lymphoblastic leukemia (ALL) is the most common cancer in children, accounting for ∼30% of all malignancies diagnosed before the age of 15 years.1 The etiology of this disease has not been fully elucidated, although known causes include ionizing radiation and congenital genetic syndromes, including neurofibromatosis type 1, Fanconi anemia, and Bloom syndrome.2 The early onset of ALL suggests an important role for inherited genetic variation in increasing risk of this cancer, and several genetic associations have already been identified through candidate gene (reviewed in Vijayakrishnan and Houlston3 ) and genome-wide association studies.4-8 In medically refractory ALL, which includes patients considered for stem cell transplantation, a spectrum of associations within the HLA complex was recently demonstrated.9 However, these findings only account for a small proportion of childhood ALL, suggesting that other genetic associations have yet to be uncovered.
In a recent study, association was reported between variation at the killer cell immunoglobulin-like receptor (KIR) gene complex and childhood ALL.10 This complex, spanning ∼150 kb at chromosome 19q13.4, is a highly variable genomic region consisting of 14 KIR genes and 2 pseudogenes.11 The KIR genes are organized head-to-tail with homologous exons and introns, peppered with varying degrees of polymorphism depending on the inhibitory or stimulatory status of the genes.12,13 This genomic structure is conducive to nonallelic homologous recombination,14,15 which has led to the formation of many different KIR gene combinations and a great amount of diversity in KIR gene repertoire between individuals12,16 and populations.17,18
KIR genes are expressed on the surface of natural killer (NK) cells and some T cells, and regulate the development and function of these cells through interaction with HLA class I ligands. Different KIR genes have varied effects on NK-cell activity: those KIR genes with long cytoplasmic tails (indicated by L in the nomenclature) transmit inhibitory signals after binding their cognate HLA ligand, and those with short tails (S) transmit activating signals.16 The expression of HLA class I alleles on the surface of cells in the body enables NK cells to recognize these as “self,” and helps them to target “non-self” entities, such as some virally infected cells and cancer cells, for lysis.19
HLA-C is the dominant KIR ligand, and all HLA-C alleles can be categorized as HLA C group 1 (C1) or group 2 (C2) based upon polymorphic residues within the α-1 extracellular domain. The C2 ligand forms a strong inhibitory interaction with the KIR2DL1 receptor, whereas C1 binds to both KIR2DL2 and KIR2DL3 receptors, which deliver intermediate and weak inhibitory signals, respectively (reviewed in Parham20 ). KIR3DL1 is an inhibitory receptor for HLA-Bw4 supertype alleles, which comprise ∼30% to 40% of HLA-B alleles.21 By contrast, HLA-Bw6 supertype alleles are not known to bind to any KIR. KIR3DL1/HLA-Bw4 interactions deliver weak or strong inhibitory signals to NK cells, depending upon the allelic polymorphisms within individual HLA-Bw4 ligands.21
There are 2 main KIR haplotypes, designated A and B, which depend upon the genes that are present.16 The A haplotype contains up to 7 expressed genes (KIR3DL3, KIR2DL3, KIR2DL1, KIR2DL4, KIR3DL1, +/−KIR2DS4, KIR3DL2) and 2 pseudogenes (KIR2DP1 and KIR3DP1). KIR B haplotypes are more variable in gene content and contain up to 14 genes, including 1 to 8 KIR genes unique to B haplotypes (KIR2DS2, KIR2DL2, KIR2DL5B, KIR2DS3, KIR3DS1, KIR2DL5A, KIR2DS5, KIR2DS1), of which the majority are stimulatory. Hence, the number of activating vs inhibitory KIR genes in an individual depends upon their KIR haplotype zygosity. The frequency of KIR haplotypes or of specific KIR genes has been associated with several diseases, including reproductive disorders,22 autoimmune diseases,23-25 susceptibility to infections,26,27 and cancer.28,29
Because association between KIR locus variation and childhood ALL was recently reported,10 but was not replicated in a subsequent analysis,30 we review and extend these genetic associations in an independent data set. Due to the important role that KIR-HLA ligand interactions may have on the development and activity of NK cells, it is essential to jointly model these loci in disease association studies. We carried out analysis on the presence or absence of all 16 KIR genes, including both inhibitory and activating KIR, along with the genotyping of their HLA-C and HLA-B ligands in 212 ALL cases and 231 healthy controls from the California Childhood Leukemia Study (CCLS). This represents the first investigation of the joint role of these highly variable immune-related loci in contributing to childhood ALL risk.
Materials and methods
Ethics statement
This study was reviewed and approved by institutional review committees at the University of California Berkeley, the California Department of Public Health, and all collaborating institutions. Written informed consent was obtained from all parent respondents. Informed consent was provided according to the Declaration of Helsinki.
Study subjects
Subjects included in this study were enrolled in the CCLS as previously described.31 Briefly, the CCLS is a continuing case-control study initiated in 1995, and includes childhood ALL cases recruited from 35 counties in Northern and Central California. One or 2 controls were selected for each case matching on age (birthdate), sex, Hispanic ethnicity, and maternal race using information from birth certificates obtained from the California Office of Vital Records. Subjects were eligible if they resided in the study area, were younger than 15 years of age at diagnosis (reference date for matched controls), had at least one English- or Spanish-speaking parent or guardian, and had no history of cancer diagnosis. Approximately 85% of eligible cases and 86% of contacted eligible controls consented to participate. Genotyping of KIR genes and their HLA ligands was carried out in 212 ALL cases and 231 controls who were born in California and had available neonatal bloodspots stored by the California Department of Public Health’s Genetic Disease Screening Program. Of these, 114 cases and 128 controls were of Hispanic ethnicity, 76 cases and 86 controls were non-Hispanic white, and 22 cases and 17 controls were non-Hispanic “other” (a mixture of African American, Asian, and other ethnicities). This subgroup of the CCLS was chosen at random from a larger case/control series, to have sufficient power to detect a significant case-control difference in KIR A/A vs B/x genotype frequency.
DNA extraction
DNA was extracted from one-eighth segments of dried blood spots collected at birth using the QIAamp DNA Micro Kit (QIAGEN). Samples from cases and controls were extracted together and subsequently diluted to 2.5 ng/μL with nuclease-free water. Samples were then randomized prior to being transferred to 96-well plates.
KIR genotyping
For genotyping the KIR loci, we used a high-throughput single nucleotide polymorphism (SNP)-based KIR genotyping assay developed using the SEQUENOM MassARRAY system and the matrix-assisted laser desorption/ionization–time of flight (MALDI-TOF) mass spectrometer platform.23,32 This assay tests for the presence or absence of 16 KIR genes (including the 2 KIR pseudogenes) and their common alleles, including the truncated allelic variants of KIR2DS4 (with 22-bp deletion, producing a protein capable of being soluble but not anchored on the cell surface) and the fully expressed variants of this stimulatory receptor.33
Briefly, 38 primer pairs are used to amplify the region surrounding the SNPs being queried. This is followed by the use of 33 homogenous mass extend primers to differentiate individual SNP patterns for the KIR genes on the MALDI-TOF platform. In this assay, multiple receptor domains are queried to identify possible recombinant loci. These assays were run using the KIR sequence alignment in the Immuno Polymorphism Database (http://www.ebi.ac.uk/ipd/kir) IPD version 2.4.0 (4/15/11). Quality control measures included blind reanalysis of 10% of samples.
HLA ligand genotyping
Genotyping of HLA-C1/C2 and HLA-Bw4/Bw6 ligand groups was performed as described in Hollenbach et al.23 Briefly, genotyping was performed using a method developed using the SEQUENOM MassARRAY system with the primer extension reactions analyzed on the MALDI-TOF mass spectrometer. The HLA-C alleles are classified as C1 or C2 KIR ligand groups, depending upon 2 amino acid (aa) positions encoded in exon 2. The C1 ligand group contains serine (AGC) at aa77 and asparagine (AAC) at aa80, while the C2 ligand group encodes asparagine (AAC) and lysine (AAA) at those positions. HLA-B alleles fall into 2 broad groups, Bw4 or Bw6, depending upon the presence of either glycine or arginine at aa83, respectively; only Bw4 supertype alleles are ligands for KIR3DL1 receptors. Amplicons containing exon 2 of HLA-C or HLA-B genes were produced using polymerase chain reaction primers designed in regions previously used for locus-specific amplification of HLA class I genes.34 Both the KIR and HLA genotyping were carried out in the Clinical Laboratory Improvement Amendments (CLIA)-approved Clinical Histocompatibility & Immunogenetics Laboratory at the Children’s Hospital & Research Center of Oakland, under the supervision of E.A.T.
Statistical analysis
Hardy-Weinberg equilibrium (HWE) analysis was carried out with 2 sets of KIR genes treated as alleles of the same locus: KIR2DL2/KIR2DL3 and KIR3DL1/KIR3DS1. Tests for HWE were performed for these KIR loci along with HLA-B and HLA-C genotypes, in all controls and also stratified by ethnicity, using contingency table testing and a standard χ2 measure, with a significance level of P = .05.
Unconditional logistic regression analyses were carried out for association tests and for generation of odds ratios (ORs) and 95% confidence intervals (CIs) using the R statistical environment (http://www.R-project.org). Several variables (sex, age at diagnosis, and ethnicity) were assessed as confounders, but were not included in the final model due to a minimal influence on risk (did not affect the OR by >5%); hence, unadjusted logistic regression models were used.
KIR haplotypes A and B were defined based on the presence or absence of specific KIR genes, as previously described.16 The SEQUENOM MALDI-TOF assay detects presence/absence but not copy number of each KIR gene, thus we were unable to distinguish between KIR B heterozygotes and homozygotes. Haplotype analysis, therefore, involved comparison of the frequency of KIR A/A vs KIR B/x genotypes, with “x” representing either the A or B haplotype (equivalent to a recessive genetic effect of KIR A). We hypothesized that KIR A/A may be associated with increased childhood ALL risk, based on the findings of Almalte et al that presence of activating KIR genes is protective against ALL.10 Power calculations were carried out (using SAS) to determine the minimal effect size this study could detect for effects of KIR A homozygosity on risk of ALL. Given a rare allele frequency of 0.30 for KIR A/A based on previous studies,30 and an α of 0.05, we were well powered (80%) to detect an effect size of OR = 1.44 in overall cases and controls, an effect size of OR = 1.61 in Hispanics only, and an effect size of OR = 1.76 in non-Hispanic whites only. These effect sizes are in line with SNP ORs from genome-wide association studies of childhood ALL.
For analysis of the frequency of each KIR gene, the presence vs absence of each gene was determined in each subject, with a P value < .003 (0.05/16 genes) regarded as statistically significant based on Bonferroni correction for multiple testing. Analysis of the total number of KIR genes between subjects involved summing the number of genes present in each subject of the 14 genes and 2 pseudogenes at the KIR locus. Neither of these analyses incorporated information on KIR gene copy number. Analysis of the number of inhibitory KIR genes included KIR2DL1, KIR2DL2, KIR2DL3, KIR2DL5, and KIR3DL1, but not the framework inhibitory KIR genes KIR2DL4, KIR3DL2, and KIR3DL3. For analysis of the activating KIR genes, in addition to KIR2DS1, KIR2DS2, KIR2DS3, KIR2DS5, and KIR3DS1, we also counted nonmutant KIR2DS4 (without the 22-bp deletion)33 as activating. Mutant KIR2DS4 (with 22-bp deletion) was counted neither as activating nor inhibitory.
Frequency of the 4 possible combinations between KIR genotype (A/A or B/x) and HLA-C alleles (C1 or C2) or HLA-B alleles (Bw4 or Bw6) was analyzed in cases and controls. Additional case-control comparisons were carried out for the frequency of known KIR gene-HLA class I ligand combinations (supplemental Table 3, available on the Blood Web site). After correction for multiple testing in both analyses, P values < .0125 (0.05/4) and < .0056 (0.05/9) were regarded as statistically significant, respectively.
In Hispanic subjects, we investigated whether ancestral differences between cases and controls may explain KIR genotype frequency. Principal components (PCs) analysis was carried out for a recent Hispanic ALL GWAS study,35 and the first 2 PCs were associated with Native American vs European ancestry and African vs European ancestry, respectively. We were, therefore, able to include these continuous ancestry-informative PCs as covariates in the logistic regression model to test whether population substructure biased analysis of the KIR A/A vs B/x genotype frequency in Hispanic case-control comparisons.
Results
Cases and controls were comparable with respect to sex, age at diagnosis (date of reference for controls), and ethnicity (Table 1).
Demographic characteristics of childhood ALL cases and controls: CCLS
| Characteristics . | n (%) . | |
|---|---|---|
| Cases, n = 212 . | Controls, n = 231 . | |
| Child’s sex | ||
| Male | 117 (55.2) | 134 (58.0) |
| Female | 95 (44.8) | 97 (42.0) |
| Mean age at diagnosis/reference, y (SE) | 4.75 (0.21) | 4.53 (0.20) |
| Child’s ethnicity | ||
| Hispanic | 114 (53.8) | 128 (55.4) |
| Non-Hispanic white | 76 (35.8) | 86 (37.2) |
| Non-Hispanic other | 22 (10.4) | 17 (7.4) |
| ALL subtype | ||
| B cell | 204 (96.2) | N/A |
| T cell | 5 (2.4) | N/A |
| Subtype unknown | 3 (1.4) | N/A |
| Characteristics . | n (%) . | |
|---|---|---|
| Cases, n = 212 . | Controls, n = 231 . | |
| Child’s sex | ||
| Male | 117 (55.2) | 134 (58.0) |
| Female | 95 (44.8) | 97 (42.0) |
| Mean age at diagnosis/reference, y (SE) | 4.75 (0.21) | 4.53 (0.20) |
| Child’s ethnicity | ||
| Hispanic | 114 (53.8) | 128 (55.4) |
| Non-Hispanic white | 76 (35.8) | 86 (37.2) |
| Non-Hispanic other | 22 (10.4) | 17 (7.4) |
| ALL subtype | ||
| B cell | 204 (96.2) | N/A |
| T cell | 5 (2.4) | N/A |
| Subtype unknown | 3 (1.4) | N/A |
N/A, not available.
KIR genes
Blind reanalysis of 10% of samples revealed complete concordance in KIR genotypes. Genotyping of all 16 KIR genes was successful in 435 of 443 subjects (98.2%): for each of the remaining 8 samples, only 1 KIR gene assay failed with the remaining 15 gene assays successful. There were no significant deviations from HWE for the KIR2DL2/KIR2DL3 or KIR3DL1/KIR3DS1 gene frequencies in either cases or controls, or when stratified by ethnicity (P > .05).
KIR haplotypes.
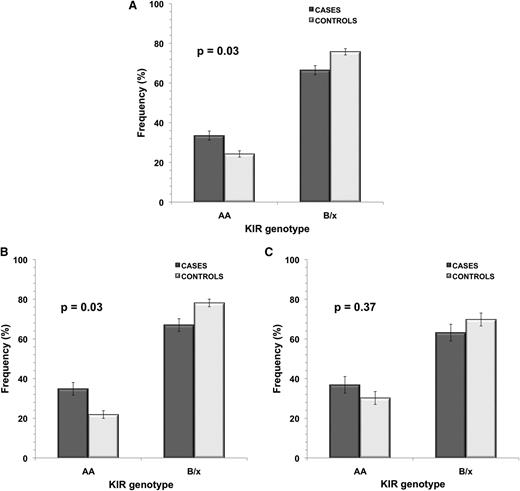
It was possible to determine KIR A haplotype homozygosity across all 443 study subjects. Frequency of KIR A/A was significantly higher in ALL cases (33.5%) compared with control subjects (24.2%), with OR = 1.57 (95% CI, 1.04-2.39; P = .03) (Figure 1 and supplemental Table 1). When subjects were stratified by ethnicity, a significant difference was seen in KIR A/A genotype frequency between Hispanic cases and controls only: 34.2% of Hispanic ALL cases were homozygous for KIR A haplotype compared with only 21.9% controls (P = .03), with OR = 1.86 (95% CI, 1.05-3.31) (Figure 1 and supplemental Table 1). In non-Hispanic whites, while there was no significant case-control difference in frequency of KIR A/A (P = .37), there was an increased frequency of this KIR genotype in cases (36.8%) compared with controls (30.2%). Restricting cases to the 204 known B-cell ALL subjects did not affect the strength of association between KIR A/A genotype and disease risk (ΔOR < 5%).
KIR genotype frequency in childhood ALL cases and controls in all subjects and in ethnic subgroups.KIR genotype frequency in childhood ALL cases vs controls is shown for (A) all subjects (cases, n = 212; controls, n = 231), (B) Hispanics only (cases, n = 114; controls, n = 128), and (C) non-Hispanic whites only (cases, n = 76; controls, n = 86). P values were calculated using unconditional logistic regression analysis between ALL cases and controls. Error bars represent SE.
KIR genotype frequency in childhood ALL cases and controls in all subjects and in ethnic subgroups.KIR genotype frequency in childhood ALL cases vs controls is shown for (A) all subjects (cases, n = 212; controls, n = 231), (B) Hispanics only (cases, n = 114; controls, n = 128), and (C) non-Hispanic whites only (cases, n = 76; controls, n = 86). P values were calculated using unconditional logistic regression analysis between ALL cases and controls. Error bars represent SE.
In Hispanic subjects, we investigated whether ancestral differences between cases and controls accounted for the significant association between KIR A/A genotype frequency and ALL risk. Adjusting for ancestry-informative PCs did not influence effect estimates for this association (ΔOR < 5%).
KIR gene frequencies.
Several genes were present at a lower frequency in cases compared with controls, although these differences did not reach statistical significance after correction for multiple testing (P > .003). Most of the genes underrepresented in cases are specific to the KIR B haplotype (supplemental Figure 1), reflecting the increased frequency of KIR A haplotype homozygotes in these individuals. A similar pattern of KIR gene frequency was seen when analyses were stratified by ethnicity, although intriguingly there was an increased frequency of KIR2DS4 and KIR3DL1 in non-Hispanic white cases compared with controls that was not seen in Hispanics (supplemental Figure 1).
Total number of KIR genes.
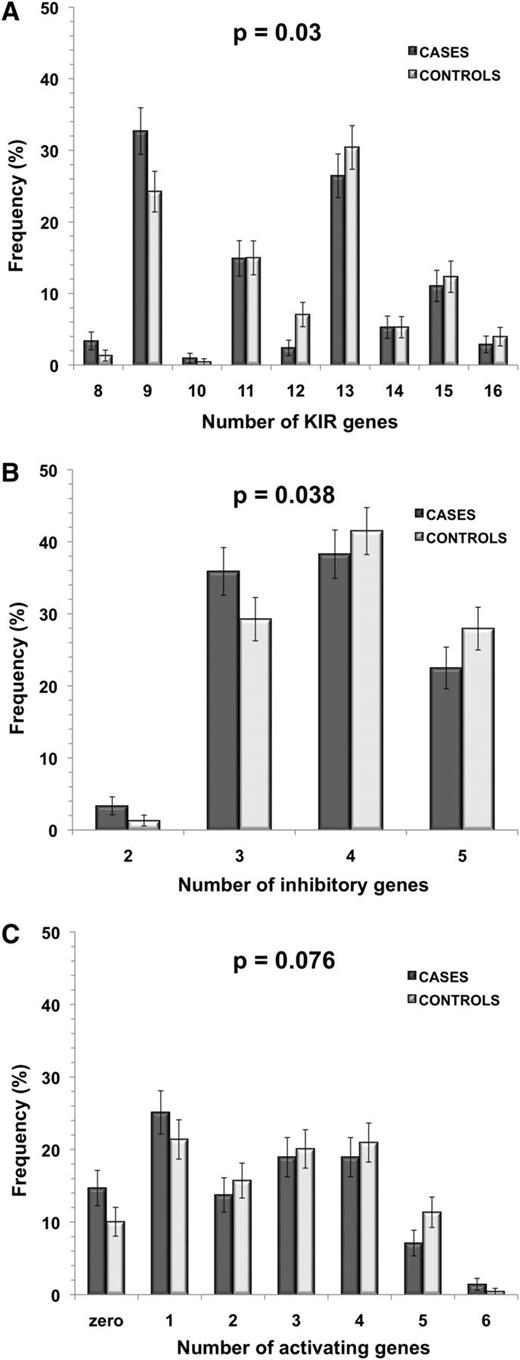
Assessment of the number of KIR genes present was carried out for 208 cases and 227 controls, as 4 cases and 4 controls were removed due to missing information. Cases had a significantly lower total number of KIR genes present than controls (P = .03; OR = 0.91; 95% CI, 0.84-0.99), with a median of 11 KIR genes present in cases and 13 in controls, reflecting the difference in KIR A/A genotype frequency (Figure 2). After assigning KIR genes to inhibitory or activating status (or neither), the total number of inhibitory KIR genes present was significantly lower in cases than controls (P = .038; OR = 0.78; 95% CI, 0.62-0.99). There was also a trend toward a lower number of activating genes present in cases compared with controls (P = .076; OR = 0.89; 95% CI, 0.80-1.01) (Figure 2). This again reflects the higher frequency of the KIR A/A genotype, in which there are fewer of both activating and inhibitory genes.
Number of KIR genes in childhood ALL cases and controls. Histograms show the number of total (A), inhibitory (B), and activating (C) KIR genes present in cases compared with controls (cases, n = 208; controls, n = 227). P values were calculated for each category using unconditional logistic regression analysis between ALL cases and controls. Error bars represent SE.
Number of KIR genes in childhood ALL cases and controls. Histograms show the number of total (A), inhibitory (B), and activating (C) KIR genes present in cases compared with controls (cases, n = 208; controls, n = 227). P values were calculated for each category using unconditional logistic regression analysis between ALL cases and controls. Error bars represent SE.
HLA ligands
Of the 443 subjects tested, 2 cases failed the HLA-C assay, 1 control failed the HLA-B assay, and 2 control samples failed both assays. These subjects were thus removed from HLA genotype analysis. In the remaining subjects, there were no significant deviations from HWE for either the HLA-C1/C2 or HLA-Bw4/Bw6 genotype frequencies in cases or controls, or when stratified by ethnicity (P > .05). There was no significant difference in the frequency of HLA-C ligand genotypes between ALL cases and controls (P = .34; OR = 1.14; 95% CI, 0.87-1.51), nor when stratified by ethnicity (supplemental Table 2).
For HLA-Bw4/Bw6, there was also no significant difference in genotype frequency between cases and controls overall (P = .17; OR = 1.23, 95% CI: 0.92-1.64). However, when analysis was stratified by ethnicity there was a significant association between ALL risk and HLA-B genotype in non-Hispanic whites (P = .02; OR = 1.74; 95% CI, 1.09-2.81), and an even stronger association between Bw4 homozygosity and increased ALL risk (P = .01; OR = 3.93; 95% CI, 1.44-12.64) (Table 2 and supplemental Figure 2). No HLA-B association was seen in Hispanics (P = .82; OR = 1.05; 95% CI, 0.69-1.61).
Frequency of HLA-B ligand genotypes in childhood ALL cases and controls in all subjects (total), Hispanics, and non-Hispanic whites: CCLS
| HLA-B genotype . | Cases (%) . | Controls (%) . | P . | OR (95% CI) . |
|---|---|---|---|---|
| Total | ||||
| n | 210 (100) | 228 (100) | ||
| Bw6/Bw6 | 95 (45.2) | 112 (49.1) | ||
| Bw4/Bw6 | 91 (43.3) | 100 (43.9) | .17 | 1.23 (0.92-1.64) |
| Bw4/Bw4 | 24 (11.4) | 16 (7.0) | ||
| Hispanics | ||||
| n | 112 (100) | 126 (100) | ||
| Bw6/Bw6 | 55 (49.1) | 66 (52.4) | ||
| Bw4/Bw6 | 52 (46.4) | 52 (41.3) | .82 | 1.05 (0.69-1.61) |
| Bw4/Bw4 | 5 (4.5) | 8 (6.4) | ||
| Non-Hispanic whites | ||||
| n | 76 (100) | 85 (100) | ||
| Bw6/Bw6 | 29 (38.2) | 42 (49.4) | ||
| Bw4/Bw6 | 32 (42.1) | 38 (44.7) | .02*† | 1.74 (1.09-2.81)* |
| Bw4/Bw4 | 15 (19.7) | 5 (5.9) | .01*‡ | 3.93 (1.44-12.64)* |
| HLA-B genotype . | Cases (%) . | Controls (%) . | P . | OR (95% CI) . |
|---|---|---|---|---|
| Total | ||||
| n | 210 (100) | 228 (100) | ||
| Bw6/Bw6 | 95 (45.2) | 112 (49.1) | ||
| Bw4/Bw6 | 91 (43.3) | 100 (43.9) | .17 | 1.23 (0.92-1.64) |
| Bw4/Bw4 | 24 (11.4) | 16 (7.0) | ||
| Hispanics | ||||
| n | 112 (100) | 126 (100) | ||
| Bw6/Bw6 | 55 (49.1) | 66 (52.4) | ||
| Bw4/Bw6 | 52 (46.4) | 52 (41.3) | .82 | 1.05 (0.69-1.61) |
| Bw4/Bw4 | 5 (4.5) | 8 (6.4) | ||
| Non-Hispanic whites | ||||
| n | 76 (100) | 85 (100) | ||
| Bw6/Bw6 | 29 (38.2) | 42 (49.4) | ||
| Bw4/Bw6 | 32 (42.1) | 38 (44.7) | .02*† | 1.74 (1.09-2.81)* |
| Bw4/Bw4 | 15 (19.7) | 5 (5.9) | .01*‡ | 3.93 (1.44-12.64)* |
P values and ORs (95% CI) calculated using unconditional logistic regression analysis.
Significant P values and ORs.
P value and OR for overall HLA-B genotype.
P value and OR for Bw4 homozygosity.
KIR-HLA ligand combinations
Frequency of the 4 different combinations between detectable KIR genotypes (A/A or B/x) with HLA-B alleles (Bw4 or Bw6) or with HLA-C alleles (C1 or C2) was determined. There was no association between KIR genotypes and HLA-B alleles (data not shown). For HLA-C, the combination of KIR A/A genotype both with C1 and with C2 were at a higher frequency in cases compared with controls, though neither reached significance after correction for multiple testing (P > .0125).
For combinations of specific inhibitory and activating KIR genes and their HLA ligands, there were no significant associations in the overall case/control analysis. However, in non-Hispanic white subjects, there was an increased frequency of the inhibitory combination between KIR3DL1 and HLA-Bw4. This case-control difference was more pronounced when limiting to subjects homozygous for the Bw4 allele, with an OR of almost 4; however, this was no longer significant after correction for multiple testing (P > .0056) (supplemental Table 3).
Discussion
In this study, we have investigated the role of KIR genes and their cognate HLA ligands in the etiology of childhood ALL, and found the KIR A/A genotype to be significantly associated with increased risk of disease. This association was more pronounced in Hispanic subjects, suggesting that an association at this locus may contribute to the well-documented higher incidence of childhood ALL in the Hispanic population.36 We also report a significant association between the HLA-Bw4 allele and risk of ALL in non-Hispanic white children, supporting a role for genetic variation at the HLA locus in the etiology of this disease. Moreover, these observations provide further evidence that functioning of the immune system in early life influences the development of childhood ALL.37
The varying effects of KIR and HLA ligand genotypes in our Hispanic and non-Hispanic white children suggest that the 2 loci may have different effects on childhood ALL susceptibility across ethnic groups. The greater case-control difference in frequency of KIR A/A genotype observed in Hispanics suggests that ALL incidence in Hispanics may be related to environmental factors that interact with KIR, for example patterns of infection, rather than simply a different allele frequency in Hispanics compared with non-Hispanic whites. The reverse might be true for the association with HLA-Bw4 in non-Hispanic whites.
Adjusting for ancestry-informative PCs in analysis of KIR genotype frequency in Hispanics revealed that significant case-control differences were not influenced by underlying ancestral differences. This is in contrast to the ARID5B, CEBPE, CDKN2A, PIP4K2A, and GATA3 ALL-risk SNPs, where risk alleles are associated with increased Native American ancestry.35,38 This discrepancy may be explained by the rapid change (via recombination and selective pressures) thought to occur at the KIR locus.18,39 Thus, the underlying KIR repertoire is likely to be independent of ancestry classifications defined by common SNP polymorphisms.
Two recent studies have reported conflicting data concerning the relationship between KIR genes and ALL risk in whites.10,30 Almalte et al10 reported a significantly reduced risk of childhood ALL with increasing numbers of activating KIR in a set of French and non-French Canadians, whereas Babor et al30 found no significant associations with individual KIR genes or KIR haplotypes in their European (mostly German) cohort. The findings of the former are supported by the higher frequency of KIR A/A genotypes in cases compared with controls in our study, as the KIR A haplotype lacks most known activating KIR genes. Furthermore, the KIR gene associations identified by Almalte et al were consistent across both B-cell and T-cell ALL, and similarly our association with KIR A/A genotype did not appear to be specific to B-cell ALL. The lack of a significant association between ALL and KIR A/A genotype frequency in non-Hispanic white subjects may support the negative findings of Babor et al, although we did see a trend for higher KIR A/A frequency in cases compared with controls.
The differences in results between the two aforementioned studies may be due to cryptic differences in the ethnicity of genotyped samples, differential accuracy of genotyping methodologies due to the strong homology between KIR genes, and/or to potential amplification biases dependent on sample quality. The latter is less of an issue for the SEQUENOM MALDI-TOF assay used in this study, which queries at least 2 domains of each KIR gene and has been shown to have high accuracy and precision (>99%), independent of sample quality.23,32 This genotyping was carried out in a CLIA-approved laboratory, and blinded repeats (10%) of our analyses demonstrated complete concordance, substantiating the assay accuracy.
An additional strength of the current study is that, in addition to KIR genes, we have also genotyped their main HLA class I ligands, alleles of HLA-C and HLA-B, enabling us to assess KIR-HLA combinations, which are pertinent to the development and activity of NK cells. Although no significant associations were detected in the overall data set with either HLA-C or HLA-B genotypes, there was a significant difference in HLA-B genotype frequency between non-Hispanic white cases and controls. Carrying 2 copies of the Bw4 allele results in an almost fourfold increased risk of childhood ALL in this ethnic group. Homozygosity for Bw4 has previously been associated with increased risk of chronic myeloid leukemia, with a trend toward increased risk of both acute myeloid leukemia and ALL,40 and we now provide evidence that this may also play a role in childhood ALL.
HLA-Bw4 binds with KIR3DL1 to deliver inhibitory signals to NK cells, and it is interesting to note that the frequency of this KIR gene was higher in non-Hispanic white cases than controls, leading to a putatively significant association between the KIR3DL1-Bw4 combination and increased ALL risk. It is possible that this inhibitory combination may enhance the ability of leukemia cells to evade immune system detection. Unlike for some solid tumors, in which downregulation of HLA class I expression is frequently observed and is a well-documented method of immune system evasion,41,42 the loss of HLA class I cell-surface expression is rare (<10%) in ALL.43 Since leukemic blasts will express HLA-Bw4 in individuals carrying this allele, the subsequent inhibition of NK cells through KIR3DL1 may lead to diminished clearance of leukemia compared with individuals lacking the Bw4 allele and/or KIR3DL1.
The present study supports a role for KIR genes in the etiology of childhood ALL; however, there are several limitations that should be considered. The finding of a Hispanic-specific association with KIR A/A genotype frequency and a non-Hispanic white–specific association with HLA-Bw4 will require further investigation in a larger group of Hispanic and non-Hispanic white patients, respectively. It is possible that the slight increase in KIR A/A frequency we observe in non-Hispanic white cases compared with controls underlies a weaker effect on ALL risk, or a comparable effect that did not reach significance due to the smaller sample size of non-Hispanic white subjects. Due to the methods used in this study, it was not possible to differentiate KIR B/B genotypes from KIR A/B ones. Therefore, it will be important to determine whether there is an additive effect of KIR A haplotypes on the risk of childhood ALL, or whether only individuals homozygous for KIR A have increased susceptibility to this disease. In addition, only presence or absence of KIR genes has been investigated and not the extensive copy number variation,44 allelic polymorphisms, and expression differences of KIR and HLA class I ligands that would further diversify the KIR-HLA interactions. Further work, therefore, is needed to elucidate the full role of the KIR and HLA ligand loci in childhood ALL.
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We gratefully acknowledge Professor Lewis Lanier and Dr James Ryan, in the Department of Immunology and Microbiology and the Department of Medicine, respectively, at University of California, San Francisco for their most helpful advice during preparation of this manuscript. We thank the families for their participation. We also thank the clinical investigators at the following collaborating hospitals for help in recruiting patients: University of California Davis Medical Center (Dr Jonathan Ducore), University of California, San Francisco (Drs Mignon Loh and Katherine Matthay), Children’s Hospital of Central California (Dr Vonda Crouse), Lucile Packard Children’s Hospital (Dr Gary Dahl), Children’s Hospital Oakland (Dr James Feusner), Kaiser Permanente Roseville (former Sacramento) (Drs Kent Jolly and Vincent Kiley), Kaiser Permanente Santa Clara (Drs Carolyn Russo, Alan Wong, and Denah Taggar), Kaiser Permanente San Francisco (Dr Kenneth Leung), and Kaiser Permanente Oakland (Drs Daniel Kronish and Stacy Month).
This work was supported by National Institutes of Health Research Program Project grants R01ES09137, P42ES004705, and P01ES018172 (P.A.B., J.L.W., C.M., and A.P.C.), grant R01CA155461 (J.L.W.), grants 5U01AI067068 and PO1CA111412 (P002051901) (E.A.T.), grant R25CA112355 (K.M.W.), and Leukemia & Lymphoma Society grant 6026-10 (J.L.W.).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Leukemia & Lymphoma Society.
Authorship
Contribution: A.J.d.S., J.L.W., E.A.T., and P.A.B. conceived and designed the study; M.B.L., S.Z., C.X., F.C., and A.J.d.S. performed the experiments; A.P.C., C.M., P.A.B., T.B.M., J.L.W., E.A.T., K.M.W., and A.J.d.S. assisted in assembling the data; K.M.W. provided data and helped with bioinformatic analyses; A.J.d.S. analyzed and interpreted the data; A.J.d.S., J.L.W., and E.A.T. wrote the manuscript; and all authors critically reviewed and edited the manuscript for intellectual content and gave final approval of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Patricia Buffler died on September 26, 2013.
Correspondence: Joseph L. Wiemels, Department of Epidemiology and Biostatistics, University of California, San Francisco, Box 0520, 1450 3rd St, San Francisco, CA 94143-0520; e-mail: joe.wiemels@ucsf.edu.