Key Points
In-depth regulome analysis of human monocyte subsets, including transcription and enhancer profiling.
Description of metabolomic differences in human monocyte subsets.
Abstract
Human blood monocytes comprise at least 3 subpopulations that differ in phenotype and function. Here, we present the first in-depth regulome analysis of human classical (CD14++CD16−), intermediate (CD14+CD16+), and nonclassical (CD14dimCD16+) monocytes. Cap analysis of gene expression adapted to Helicos single-molecule sequencing was used to map transcription start sites throughout the genome in all 3 subsets. In addition, global maps of H3K4me1 and H3K27ac deposition were generated for classical and nonclassical monocytes defining enhanceosomes of the 2 major subsets. We identified differential regulatory elements (including promoters and putative enhancers) that were associated with subset-specific motif signatures corresponding to different transcription factor activities and exemplarily validated novel downstream enhancer elements at the CD14 locus. In addition to known subset-specific features, pathway analysis revealed marked differences in metabolic gene signatures. Whereas classical monocytes expressed higher levels of genes involved in carbohydrate metabolism, priming them for anaerobic energy production, nonclassical monocytes expressed higher levels of oxidative pathway components and showed a higher mitochondrial routine activity. Our findings describe promoter/enhancer landscapes and provide novel insights into the specific biology of human monocyte subsets.
Introduction
Human blood monocytes are mobile phagocytes that are able to differentiate into an array of functionally different cell types, including macrophages, myeloid dendritic cells, and osteoclasts.1,2 In humans, 3 major subpopulations of monocytes are distinguished by their differential surface expression of the coreceptor for lipopolysaccharide CD14 and the FcγIII receptor CD16.3 “Classical” monocytes, which express high levels of CD14 and no CD16 (CD14++CD16−), are the largest subgroup of circulating monocytes. “Nonclassical” monocytes have low or absent CD14 but express CD16 (CD14dimCD16+),3 and “intermediate” monocytes express both markers. Monocyte subsets have been ascribed distinct functions and fates in both humans and mice (where they are distinguished by expression of the surface marker Ly6C).4-7 Comparative array profiling has been used as evidence to support orthology between the human and mouse subsets (CD14hi = Ly6Chi), but many genes show species-specific expression profiles.8 The ontogenetic relationship between monocyte subsets is a subject of debate. Based on fluorescence-activated cell sorter profiling, gene expression, transplantation, and antibody-inhibition studies, several studies favor a linear differentiation model whereby classical monocytes convert to more mature nonclassical monocytes over time.1,6,9 More recent work in the murine system challenges the linear model and proposes that the nonclassical subset has an independent origin from classical monocytes: Ly6C− monocytes, the “murine counterpart” of the nonclassical human subset, depend on the transcription factor NR4A1 that apparently controls the differentiation of this subset from immature proliferating precursors in the bone marrow.10
Previous microarray studies4,8,11-13 and a recent serial analysis of gene-expression technology profiling14 have identified genes that distinguish monocyte subsets but have not addressed the mechanisms of transcriptional regulation. Here, we analyzed gene expression as well as promoter and enhancer usage by cap analysis of gene expression adapted to Helicos single-molecule sequencing (HeliScopeCAGE)15,16 alongside genome-wide epigenetic profiling. Our study provides a detailed picture of the cis-regulatory landscape of human monocyte subsets and identifies novel biological differences between classical and nonclassical subpopulations. This work is part of the Functional Annotation of Mammalian Genome 5 (FANTOM5) project. Data downloads, genomic tools, and copublished manuscripts are summarized online at http://fantom.gsc.riken.jp/5/.
Materials and methods
Cells
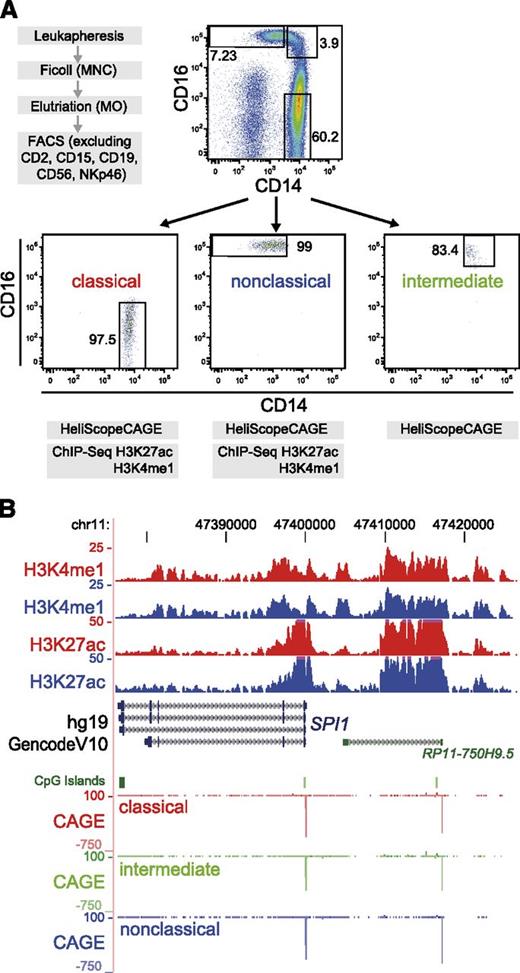
Peripheral blood monocytes were separated by leukapheresis of healthy donors, density-gradient centrifugation over Ficoll/Hypaque, and subsequent countercurrent centrifugal elutriation as previously described.17,18 Collection of blood cells from healthy donors was performed in compliance with the Declaration of Helsinki. All donors signed an informed consent. The procedure was approved by the local ethics committee (reference number 92-1782 and 09/066c). The monocyte-enriched cell fraction was further subdivided by fluorescence-activated cell sorting (FACS) (FACSAria I; BD Biosciences, Heidelberg, Germany) after staining with CD2-PE (RPA-2.10), CD15-PE (HI98), CD19-PE (4G7), CD56-PE (B159), and NKp46-PE (9E2), to exclude remaining lymphocytes and granulocytes, as well as CD14-V450 (MΦP9) and CD16-Alexa647 (3G8) antibodies (all from BD Biosciences). Monocytes were sorted into CD14++CD16− (classical subset), CD14+CD16+(intermediate subset) and CD14dimCD16+ cells (nonclassical subset) (Figure 1).19 THP-1 (human monocytic cell line) and Jurkat cells (human T-cell leukemia) were cultured as previously described.17,20
Experimental setup and regulome profile of the human SPI1 locus. (A) Isolation scheme for monocyte subsets, sort gates for the FACS purification, and reanalysis of the 3 CD14/CD16-stained monocyte subsets are shown (1 representative out of 3 independent experiments). (B) Genome browser tracks covering the human SPI1 locus of available epigenetic (H3K4me1 and H3K27ac) and HeliScopeCAGE data (coloring indicates monocyte subtypes: classical, red; intermediate, green; nonclassical, blue). All 3 subsets equally express the myeloid master regulator PU.1, and the 2 major subsets show strikingly similar epigenetic profiles across the locus. The second CAGE peak at right corresponds to the position of the conserved upstream enhancer, well characterized in mice.19
Experimental setup and regulome profile of the human SPI1 locus. (A) Isolation scheme for monocyte subsets, sort gates for the FACS purification, and reanalysis of the 3 CD14/CD16-stained monocyte subsets are shown (1 representative out of 3 independent experiments). (B) Genome browser tracks covering the human SPI1 locus of available epigenetic (H3K4me1 and H3K27ac) and HeliScopeCAGE data (coloring indicates monocyte subtypes: classical, red; intermediate, green; nonclassical, blue). All 3 subsets equally express the myeloid master regulator PU.1, and the 2 major subsets show strikingly similar epigenetic profiles across the locus. The second CAGE peak at right corresponds to the position of the conserved upstream enhancer, well characterized in mice.19
RNA preparation
RNA for cap analysis of gene expression (CAGE) sequencing and quantitative reverse-transcription polymerase chain reaction (qRT-PCR) was isolated using the miRNeasy RNA isolation kit (Qiagen, Hilden, Germany). For qRT-PCR, DNA was removed completely with the RNase-free DNase set (Qiagen). RNA quality was checked with the Bioanalyzer (Agilent Technologies, Böblingen, Germany).
Quantitative RT-PCR
Total RNA was transcribed into complementary DNA (cDNA) with reverse-transcriptase RNase H (−) point mutation (Promega, Madison, WI) and analyzed on an Eppendorf Realplex4 S cycler (Eppendorf International). Expression levels of individual genes were normalized to ACTB expression. Primer sequences are listed in supplemental Table 1 (available on the Blood Web site).
HeliScopeCAGE sequencing and data analysis
Single-molecule HeliScopeCAGE16 data were prepared as described elsewhere.15 Normalization of tag libraries was done using the common power-law distribution approach developed by Balwierz et al.21 Expression data for annotated coding or noncoding genes (according to GencodeV10) was extracted by collecting normalized tag counts in regions −500 to +200 relative to all annotated transcription start sites (TSSs) associated with a single gene ID. Digital gene expression analysis of normalized data was performed using edgeR.22 Clustering of significantly differentially expressed genes (5% false-discovery rate cutoff) was done using Cluster 3.0.23 Functional annotation of differentially regulated genes (classical/nonclassical: P ≤ .05 for pairwise comparison; intermediate: expression higher than both other subsets and P ≤ .05 for at least 1 pairwise comparison) was done using DAVID tools.24
5′-RACE PCR
Rapid amplification of cDNA ends (RACE) was performed with the SMARTer RACE cDNA amplification kit (Clontech, Saint-Germain-en-Laye, France) and the Advantage 2 polymerase system (Clontech) using either a single gene-specific primer or a second nested primer (ngsp1) when required. Single bands were cloned and sequenced (Life Technologies, Regensburg, Germany). Primer sequences are listed in supplemental Table 1.
ChIP-sequencing and data analysis
Chromatin was obtained from classical and nonclassical monocytes of 2 healthy donors. Chromatin immunoprecipitation (ChIP), library construction, mapping and peak calling were done essentially as described elsewhere.18 For histone vs CAGE data comparisons, sequence tags from different donors were combined and tag counts were normalized to 107 tags to determine differentially marked regions. Regions showing at least 3-fold tag count differences between cell types were considered different. ChIP-sequencing data have been deposited with the National Center for Biotechnical Information’s Gene Expression Omnibus database (accession code GSE40502) and are listed in supplemental Table 2. UCSC Genome Browser track hub data for the entire data set can be found at http://www.ag-rehli.de/NGSdata.htm.
De novo motif analyses
Enriched sequence motifs were de novo extracted from regions surrounding differentially expressed CAGE clusters, differentially marked H3K27ac peaks, and differentially marked promoter-distal H3K4me1 peaks using HOMER25 as detailed in supplemental Methods.
Network analysis
We inferred regulatory inputs of genes differentially expressed in the 3 monocyte subsets samples by applying a feature-selection approach similar to the one used in the program Genie326 as outlined in supplemental Methods. The network was constructed from the top 50 transcription factors defined by the number of target genes in each monocyte subset (listed in supplemental Table 3) using the STRING 9.0 database.27
Transient DNA transfections
The human CD14 promoter and putative enhancer regions were amplified from monocyte genomic DNA using PCR primers listed in supplemental Table 1. PCR fragments were cloned upstream of the human CD14 promoter in the pGL4.10 vector (Promega) and sequenced for validation. THP-1 and Jurkat cells were transfected and treated with phorbolmyristate acetate as described elsewhere.20,28 Transfections correspond to at least 3 independent experiments measured in triplicate.
High-resolution respirometry
Activity of the respiratory system was analyzed in a 2-channel titration injection respirometer (Oxygraph-2k; Oroboros, Innsbruck, Austria) at 37°C. After FACS, cells were centrifuged, resuspended in mitochondrial medium MiR05,29 and transferred to the oxygraph chambers. After a stabilization phase of 15 to 20 minutes, routine respiration of intact cells was measured. Residual oxygen consumption was determined after the addition of rotenone (complex I inhibitor) and antimycin A (complex III inhibitor). Complex IV activity was determined in digitonin-permeabilized (16.2 µM) cells by using TMPD (N,N,N′,N′-tetramethyl-p-phenylenediamine dihydrochloride) as substrate and ascorbate to keep TMPD in a reduced state.
Citrate synthase assay
Specific activity of the mitochondrial marker citrate synthase (CS) was measured photometrically at 412 nm using an established protocol.30 Respiration rates were calculated as the time derivative of oxygen concentration (pmol/[s*mio cells]) and normalized to CS activity (pmol/[s*mio cells]).
MitoTracker staining
Enrichment of monocytes was done by depleting lymphocytes labeled with the lineage panel as described above. The enriched fraction was stained with MitoTracker Green FM (25 nM, Invitrogen) for 2 hours under cell-culture conditions. Cells were washed, CD14/16 stained, and analyzed by flow cytometry.
Western analysis
Western blotting was performed using whole-cell extracts as described previously.17 Details and antibodies used are provided in supplemental Methods.
Results
To study promoter and enhancer usage in monocyte subsets, we devised a purification strategy based on leukapheresis, elutriation, and subsequent FACS, which is outlined in Figure 1A. RNA was isolated for all 3 subpopulations, and chromatin was additionally prepared for classical and nonclassical subsets. As part of the FANTOM5 expression atlas,15 RNA was subjected to HeliScopeCAGE sequencing, a method that avoids second-strand synthesis, ligation, digestion, and PCR to provide an unbiased, digital readout for gene expression and promoter usage.16 To extend the analysis of regulatory sites to putative enhancers, we also carried out ChIP sequencing for 2 histone marks, namely H3K4me1 and H3K27ac, that were previously associated with enhancers.30-34 H3K27 is a major substrate for the coactivators p300 and CBP and its acetylation marks active enhancers, whereas H3K4me1 is generally associated with distal regulatory elements, including poised enhancers (reviewed in Calo and Wysocka35 ).
The SPI1 locus encoding the myeloid and B-cell–specific transcription factor PU.1 is shown as an example that clearly characterizes all subsets as monocytes (Figure 1B). To allow easy viewing of this resource, we created a UCSC Genome Browser track hub that is available at http://www.ag-rehli.de/NGSdata.htm.
One major advantage of CAGE is the exact determination of TSSs, which allows the characterization of upstream regulatory sequences. The current annotation of TSSs often differs from CAGE data. Of the 36K CAGE TSSs that show robust expression (≥1 tags per million [TPM]) in any of the monocyte subsets, 41% and 29% were not located in the vicinity (±500 bp) of TSSs annotated in RefSeq or GencodeV10 collections, respectively. 5′-RACE PCR for examples where annotated and HeliScopeCAGE TSS differed confirmed the CAGE data in all 4 cases (supplemental Figure 1), corroborating that CAGE accurately determined monocyte TSSs. Similar results were obtained in T cells36 and were also extensively validated in a previous study of THP-1 differentiation.37
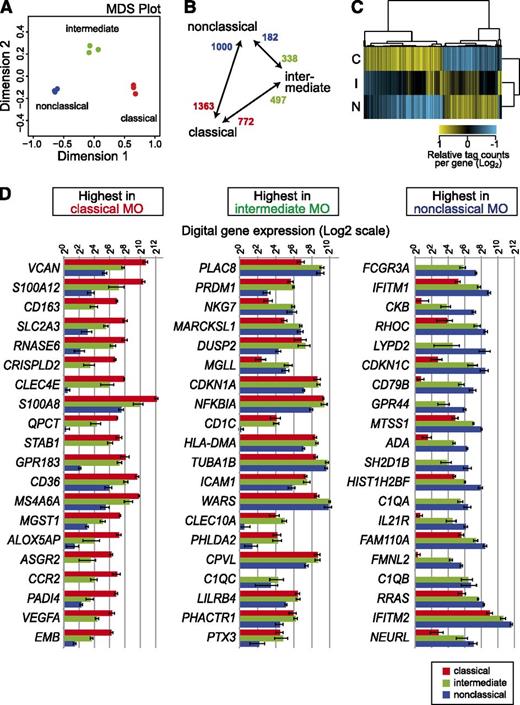
A total of 10 249 GeneCodeV10 protein-coding genes were associated with at least 1 TPM in monocytes with correlation coefficients for the level of expression ranging between 0.96 and 0.98 for replicate pairs (a correlation matrix is shown in supplemental Figure 2). The monocyte subsets were clearly separated in a multidimensional scaling plot (Figure 2A). Two-dimensional hierarchical clustering of differentially expressed genes (5% false-discovery rate; for numbers, see Figure 2B) indicates that the intermediate subset represents a true intermediate in the majority of cases (Figure 2C). In line with previous profiling studies,5,14 a relatively small proportion of genes was specifically upregulated in the intermediate set. The 2 major subsets were found to express most genes that were previously attributed to these monocytes. These included CD14, CCR2, CD163, VCAN, S100A8, A9, and A12 for the classical subset and FCGR3A (CD16), CDKN1C, TCF7L2, C1QA, QB, and QC for the nonclassical subset. Fewer genes were specific for the intermediate subset, including several HLA genes as well as EGR1 and EGR2 genes. The top 20 differentially expressed genes (ranked by q-value) in each monocyte subset are shown in Figure 2D, and the entire data set for coding genes is available in supplemental Table 4. Interestingly, 2 of the top 20 genes that distinguish nonclassical monocytes are IFITM1 and IFITM2, and the expression of the neighboring IFITM3 locus (supplemental Table 4) shows the same pattern. All 3 proteins have been implicated in resistance to multiple viruses.38 The expression data suggest some form of locus control and also imply a role for nonclassical monocytes in innate antiviral defense.
HeliScopeCAGE-based digital expression analysis. For digital gene expression analysis, tag counts were collected within −500 to +200 bp of GencodeV10 annotated coding genes as outlined in “Materials and methods.” (A) The multidimensional scaling (MDS) plot for replicate HeliScopeCAGE-based digital expression data shows that samples are separated by at least 1 dimension. (B) Numbers of differentially expressed genes in pairwise comparisons of subset samples are given. (C) Two-dimensional hierarchical clustering of 2600 differentially expressed coding genes (normalized to the geometric mean, log2 transformed). C, classical; I, intermediate; N, nonclassical monocyte subset. (D) Digital gene expression data for the top 20 monocyte (MO) subset-specific genes.
HeliScopeCAGE-based digital expression analysis. For digital gene expression analysis, tag counts were collected within −500 to +200 bp of GencodeV10 annotated coding genes as outlined in “Materials and methods.” (A) The multidimensional scaling (MDS) plot for replicate HeliScopeCAGE-based digital expression data shows that samples are separated by at least 1 dimension. (B) Numbers of differentially expressed genes in pairwise comparisons of subset samples are given. (C) Two-dimensional hierarchical clustering of 2600 differentially expressed coding genes (normalized to the geometric mean, log2 transformed). C, classical; I, intermediate; N, nonclassical monocyte subset. (D) Digital gene expression data for the top 20 monocyte (MO) subset-specific genes.
Similar analyses were also performed for known noncoding genes. In total, 913 noncoding genes with >1 TPM were detected. The set of differentially expressed noncoding RNA genes was considerably smaller (99 genes in total). Expression counts for the top 20 differentially expressed noncoding genes in each monocyte subset are shown in supplemental Figure 3. Of note, the subset-specific noncoding RNAs also included the precursor RNA (CTC-231O11.1) for miR-146a, which was recently associated with CD16+ monocytes.39 Data for noncoding genes are available in supplemental Table 5.
At promoter-distal regions, the histone modifications H3K4me1 and H3K27ac are thought to mark “poised” (H3K4me1 alone) or “active” enhancers (H3K4me1 and H3K27ac).30-34 As shown in supplemental Figure 4, in monocyte chromatin the average distribution of both epigenetic marks detected relative to CAGE-cluster centers almost perfectly correlated with CAGE expression levels. The chromatin data are in line with earlier ChIP-sequencing data on monocytes18 and, for example, confirmed a transcribed enhancer in the second intron of the CSF1R locus (FIRE).40 The corresponding CAGE data also confirmed that the enhancer (FIRE) initiates antisense transcripts; the importance of antisense transcription has recently been confirmed experimentally in the mouse.41
To globally link enhancers with candidate target promoters, we analyzed CAGE clusters surrounding enhancer regions as well as their expression level (represented by CAGE tag counts). As shown in the bubble plot representations in Figure 3A for H3K27ac (in supplemental Figure 5 for H3K4me1), subset-specific enhancers were significantly associated with higher tag counts in neighboring CAGE clusters.
Monocyte subset-specific active enhancer signatures. (A) Bubble-plot representation of CAGE-TSS activity around subset-specific enhancer candidate regions showing at least 3-fold different H3K27ac signals. The bubble plots encode 3 quantitative parameters per CAGE cluster: distance from the putative enhancer, log10 of fold change in CAGE cluster tag count between classical and nonclassical monocytes (y-axis), and the absolute CAGE cluster tag count of the monocyte subset with the highest expression level (bubble diameter). There is a clear bias for the putative enhancer elements to associate with CAGE clusters upregulated in the corresponding cell type (P < .001, Wilcoxon signed-rank test). (B) Putative subset-specific enhancer region of the human CD14 locus. On top, UCSC Genome Browser tracks covering the entire CD14 locus are shown including epigenetic data (H3K4me1 and H3K27ac), positions of transcription factor binding sites for PU.1 and C/EBPβ in total monocytes,18 positions of bidirectional enhancers identified from the FANTOM5 expression atlas,44 HeliScopeCAGE data (monocyte subsets are indicated by coloring: classical, red; intermediate, green; nonclassical, blue), GencodeV10 gene annotation, 5′-RACE–based annotation of a novel TMOC6 transcript, and positions of genomic intervals that were used for enhancer reporter assays. The bottom panel shows relative luciferase activities (enhancer construct/CD14 promoter only [control]) of individual enhancer constructs in a monocytic (THP-1) and a T-cell (Jurkat) cell line. A myeloid-specific set of CD14 enhancers was detected between regions 6 and 8.
Monocyte subset-specific active enhancer signatures. (A) Bubble-plot representation of CAGE-TSS activity around subset-specific enhancer candidate regions showing at least 3-fold different H3K27ac signals. The bubble plots encode 3 quantitative parameters per CAGE cluster: distance from the putative enhancer, log10 of fold change in CAGE cluster tag count between classical and nonclassical monocytes (y-axis), and the absolute CAGE cluster tag count of the monocyte subset with the highest expression level (bubble diameter). There is a clear bias for the putative enhancer elements to associate with CAGE clusters upregulated in the corresponding cell type (P < .001, Wilcoxon signed-rank test). (B) Putative subset-specific enhancer region of the human CD14 locus. On top, UCSC Genome Browser tracks covering the entire CD14 locus are shown including epigenetic data (H3K4me1 and H3K27ac), positions of transcription factor binding sites for PU.1 and C/EBPβ in total monocytes,18 positions of bidirectional enhancers identified from the FANTOM5 expression atlas,44 HeliScopeCAGE data (monocyte subsets are indicated by coloring: classical, red; intermediate, green; nonclassical, blue), GencodeV10 gene annotation, 5′-RACE–based annotation of a novel TMOC6 transcript, and positions of genomic intervals that were used for enhancer reporter assays. The bottom panel shows relative luciferase activities (enhancer construct/CD14 promoter only [control]) of individual enhancer constructs in a monocytic (THP-1) and a T-cell (Jurkat) cell line. A myeloid-specific set of CD14 enhancers was detected between regions 6 and 8.
On this basis, we sought an explanation for the differential expression of CD14 in classical monocytes. Studies in transgenic mice by Zhang and coworkers established that a region of 80 kb surrounding the human CD14 gene is sufficient to direct its monocyte-specific expression,42 whereas smaller constructs recapitulated human liver expression but failed to direct monocyte-specific expression.43 The genomic interval downstream of the CD14 gene contained a number of H3K27ac-marked sites specific for classical, CD14-expressing monocytes (regions 3, 4, 6-8, and 10; Figure 3B). These sites frequently overlapped with bidirectional enhancers identified using the FANTOM5 expression atlas44 as well as binding sites for PU.1 or C/EBPβ in total monocytes,18 which are key factors in establishing distal regulatory sites in these cells.18,25 As shown in the bottom panel of Figure 3B, control regions 1, 2, 5, and 9 as well as the 2 putative promoter regions (3, APBB3; 4, TMCO6) and the region immediately downstream of CD14 show little or no enhancer activity. Three candidate regions (6-8) ∼20 kb downstream of CD14 clearly enhanced promoter activity specifically in the monocytic cell line THP1, but not in CD14-negative Jurkat T cells. Hence, these sites represent distal regulatory elements critical for the cell-type–specific expression of this locus.
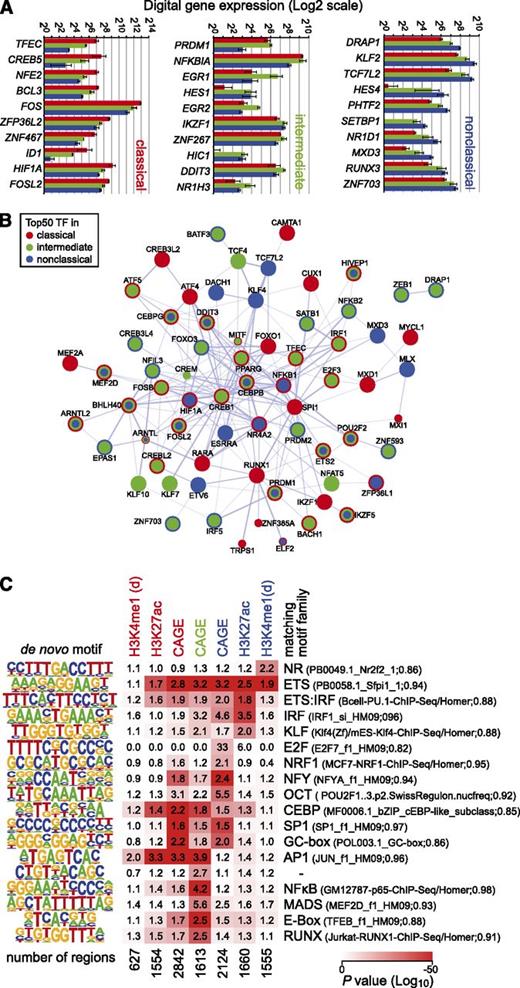
The monocyte subsets can be distinguished by gene expression of transcription factors; the top 10 differentially expressed transcription factor genes in each monocyte subset are shown in Figure 4A. The data confirm the differential expression of the myeloid transcription factor TFEC,8 but NR4A110 was expressed equally low in all subsets.
Transcription factor expression and motif enrichment. (A) Digital gene expression (DGE) data for the top 10 monocyte subset-specific transcription factor genes. (B) Combined STRING-based network (confidence view) of the top transcription factors predicted to have the strongest regulatory input in either subset. Coloring of nodes indicates the presence of the transcription factor in the top list of the respective monocyte subset. (C) Enrichment of motifs that were de novo generated from subset-specific CAGE clusters as well as regions showing subset-specific H3K4me1 or H3K27ac deposition as outlined in supplemental Methods. P values for motif enrichment were calculated using the hypergeometric test relative to the distribution in a random, GC-normalized background set. Data are presented as a heatmap (log10 scaled), where red coloring indicates significant motif enrichment in the given sets of subset-specific regions. Numbers in boxes represent relative changes in motif enrichment. De novo–derived motifs were compared with known motifs, and the best-matching motif families are given on the right. The total number of regions per set is given below each heatmap column. For H3K4me1, only promoter-distal regions (d) were considered.
Transcription factor expression and motif enrichment. (A) Digital gene expression (DGE) data for the top 10 monocyte subset-specific transcription factor genes. (B) Combined STRING-based network (confidence view) of the top transcription factors predicted to have the strongest regulatory input in either subset. Coloring of nodes indicates the presence of the transcription factor in the top list of the respective monocyte subset. (C) Enrichment of motifs that were de novo generated from subset-specific CAGE clusters as well as regions showing subset-specific H3K4me1 or H3K27ac deposition as outlined in supplemental Methods. P values for motif enrichment were calculated using the hypergeometric test relative to the distribution in a random, GC-normalized background set. Data are presented as a heatmap (log10 scaled), where red coloring indicates significant motif enrichment in the given sets of subset-specific regions. Numbers in boxes represent relative changes in motif enrichment. De novo–derived motifs were compared with known motifs, and the best-matching motif families are given on the right. The total number of regions per set is given below each heatmap column. For H3K4me1, only promoter-distal regions (d) were considered.
To identify regulators that are most important in describing the expression pattern of subset-specific target genes, we used a regularized random forest-based regression analysis.26 The combination of networks (based on the STRING database of known and predicted protein interactions27 ) of the top transcription factors relevant for each monocyte subset (Figure 4B; supplemental Table 3) highlights the presence of subset-specific regulatory inputs (single colored nodes in Figure 4B). Several regulators included in this network were also differentially expressed between subsets. To further link differentially expressed transcription factors to candidate targets, we also used de novo motif detection within subset-enriched promoters and enhancers. Subset-specific CAGE clusters as well as regions showing subset-specific H3K4me1 or H3K27ac deposition contained sequence motifs that were highly enriched in individual subsets. Figure 4C shows the compiled and nonredundant set of de novo–derived sequence motifs together with their enrichment over random background sequences in subset-specific regulatory regions. In line with previous findings,18,25 the motif signature of classical monocytes is dominated by AP-1 and CEBP (both CAGE TSS and putative enhancers) as well as 2 GC-rich motifs that were exclusively enriched in CAGE clusters. Conversely, nuclear factor κB, E-box, and MEF2 motifs were enriched in the intermediate subset, whereas the nonclassical signature included promoter-enriched motifs like E2F, NRF1, and OCT as well as promoter-distal ETS, IRF, KLF, and nuclear receptor motifs. Several of the signature motifs correspond to regulated candidate binding factors, including FOS (AP1 motif), KLF2 (KLF motif), and E2F1 (E2F motif), or were also part of the subset-specific networks (like RUNX1 [RUNX motif] in classical monocytes, BACH1 [AP1 motif] in classical and intermediate monocytes, and KLF4 [KLF motif] in the nonclassical subset). De novo motif finding results are also in line with TF motif networks provided in the general FANTOM5 study (http://fantom.gsc.riken.jp/5/sstar/).15 Although we do observe matching pairs of motifs and corresponding transcription factors, it is clear that messenger RNA expression alone is not equivalent for binding or activity (which may be regulated by the presence of ligands, posttranscriptionally or posttranslationally), and hence not all (differentially) expressed transcription factors are expected to leave a motif fingerprint under nonstimulated steady-state conditions (eg, HIF1A or TCF7L2). On the other hand, de novo motifs also included a motif resembling the PU.1 consensus sequence that is enriched in all subset-specific promoter and active enhancer sets, suggesting that a factor (like PU.1), which is not differentially expressed, may still participate in subset specific regulatory events.
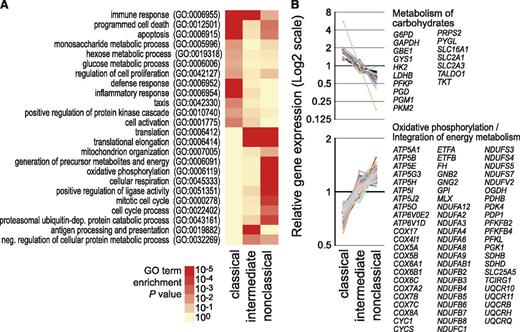
As noted above, monocyte subsets have been ascribed distinct functions. As shown in Figure 5A, analysis of the differentially expressed genes for enriched Gene Ontology (GO) terms and pathways related to metabolic processes confirmed known associations of classical monocytes with lysosome-associated genes and nucleotide oligomerization domain and Toll-like receptor pathways (supplemental Figure 6). We also noted the upregulation of genes involved in carbohydrate metabolism, in particular genes involved in the glycolytic and the pentose phosphate pathway. This included glycolytic pathway genes like glucose transporter GLUT3 (SLC2A3), hexokinase 2 (HK2), glyceraldehyde 3-phosphate dehydrogenase (GAPDH), phosphofructokinase (PFKP), and lactate dehydrogenase (LDHB) and pentose phosphate pathway genes like the 2 genes encoding reduced NAD phosphate (NADPH)-generating enzymes glucose 6-phosphate dehydrogenase (G6PD) and 6-phosphogluconate dehydrogenase (PGD) as well as transketolase (TKT) and transaldolase (TALDO1). In line with previous work,13,14 the intermediate subset was characterized by the upregulation of genes associated with antigen processing and presentation. The nonclassical set, however, was associated with the oxidative phosphorylation pathway (see Figure 5B) and protein metabolism. In particular, a large number of genes encoding components of complexes I, II, III, and V of the mitochondrial respiratory chain showed consistently higher expression in the nonclassical monocyte subset (Figure 5B). The majority of expression differences observed by CAGE profiling were validated by qRT-PCR; however, the normalization to ACTB abolished most differences in the respiratory chain genes (supplemental Figure 7A). A subset of enzymes, including HK2, SLC2A3, and LDHB, was also analyzed on protein level, confirming the metabolic bias in classical monocytes (supplemental Figure 7B).
HeliScopeCAGE-based GO and pathway analyses. (A) Heatmap (log10 scaled) of significance values for GO enrichment terms related to biological processes. Enrichment for GO terms was calculated for subset-specific gene sets using DAVID tools.24 P values for enrichment of each GO term were calculated using the hypergeometric test and corrected for multiple hypothesis testing using the Benjamini-Hochberg procedure. Intensity of the red coloring indicates the significance of term enrichment. Only generic terms are given. (B) Relative HeliScopeCAGE-based gene expression levels of genes in enriched pathways. Symbols of subset-specific genes are provided for each pathway. Additional examples, including lysozyme, nucleotide oligomerization domain-like/Toll-like receptor–like receptor signaling pathway, antigen processing and presentation, ribosome, and protein metabolism are shown in supplemental Figure 6. Notably, the observed differences were independent of the normalization method and also observed using normalization to a fixed tag count (TPM) or relative log expression normalization (as provided by EdgeR) of raw tag counts.
HeliScopeCAGE-based GO and pathway analyses. (A) Heatmap (log10 scaled) of significance values for GO enrichment terms related to biological processes. Enrichment for GO terms was calculated for subset-specific gene sets using DAVID tools.24 P values for enrichment of each GO term were calculated using the hypergeometric test and corrected for multiple hypothesis testing using the Benjamini-Hochberg procedure. Intensity of the red coloring indicates the significance of term enrichment. Only generic terms are given. (B) Relative HeliScopeCAGE-based gene expression levels of genes in enriched pathways. Symbols of subset-specific genes are provided for each pathway. Additional examples, including lysozyme, nucleotide oligomerization domain-like/Toll-like receptor–like receptor signaling pathway, antigen processing and presentation, ribosome, and protein metabolism are shown in supplemental Figure 6. Notably, the observed differences were independent of the normalization method and also observed using normalization to a fixed tag count (TPM) or relative log expression normalization (as provided by EdgeR) of raw tag counts.
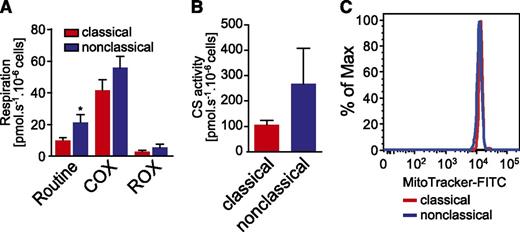
To link these observations to function, we measured mitochondrial respiratory activity in the 2 major subsets. Basal respiration was significantly elevated in nonclassical monocytes from all donors (Figure 6A). Residual oxygen consumption and complex IV (COX) activity showed the same tendency (Figure 6A), although there was greater variability between the donors. In addition, we determined CS activity, which is described as a marker enzyme for mitochondrial matrix and content.45-48 All donors showed increased CS activity in nonclassical monocytes compared with classical monocytes (Figure 6B). Normalizing routine respiration and COX activity to CS activity abolished the differences on cellular level, proposing that alterations are not restricted to respiration (supplemental Figure 7C). In addition, MitoTracker staining revealed no differences between the 2 subsets, indicating that the activities, but not the content, of mitochondria are different between the 2 subsets (Figure 6C).
Comparative metabolic analyses of classical vs nonclassical monocytes. (A) Basal mitochondrial respiratory activity (Routine), complex IV activity (COX), and residual oxygen consumption (ROX) were determined by high-resolution respirometry in both classical and nonclassical monocyte populations. Routine and COX activity were corrected for ROX respiration, because this oxygen consumption is not related to the mitochondrial respiratory system. All parameters were increased and the elevation of routine respiration was significant (P ≤ .05, Wilcoxon matched-pairs test) in nonclassical monocytes. Shown are arithmetic means and standard errors of the mean (n = 6 for routine respiration, n = 5 for COX and ROX). (B) The activity of CS, a mitochondrial matrix marker enzyme, is increased in nonclassical monocytes. Shown are arithmetic means and standard error of the mean (n = 5). The difference was not significant in the Wilcoxon matched-pairs test. (C) Mitochondrial content, determined by MitoTracker staining and analyzed by flow cytometry, was not different between the 2 subpopulations. Shown is 1 representative experiment of n = 3. FITC, fluorescein isothiocyanate.
Comparative metabolic analyses of classical vs nonclassical monocytes. (A) Basal mitochondrial respiratory activity (Routine), complex IV activity (COX), and residual oxygen consumption (ROX) were determined by high-resolution respirometry in both classical and nonclassical monocyte populations. Routine and COX activity were corrected for ROX respiration, because this oxygen consumption is not related to the mitochondrial respiratory system. All parameters were increased and the elevation of routine respiration was significant (P ≤ .05, Wilcoxon matched-pairs test) in nonclassical monocytes. Shown are arithmetic means and standard errors of the mean (n = 6 for routine respiration, n = 5 for COX and ROX). (B) The activity of CS, a mitochondrial matrix marker enzyme, is increased in nonclassical monocytes. Shown are arithmetic means and standard error of the mean (n = 5). The difference was not significant in the Wilcoxon matched-pairs test. (C) Mitochondrial content, determined by MitoTracker staining and analyzed by flow cytometry, was not different between the 2 subpopulations. Shown is 1 representative experiment of n = 3. FITC, fluorescein isothiocyanate.
Taken together, the above functional studies suggest that classical and nonclassical monocyte subsets show major differences in energy metabolism.
Discussion
Initial evidence for monocyte subpopulations based on cell size and density from the early 1980s (reviewed in Auffray et al49 ) was later corroborated by the identification of cell-surface markers that distinguished monocyte subpopulations.7 The present study extends earlier work on the transcriptional landscape that distinguishes these cells. It represents a unique resource for monocyte biologists, including genome-wide CAGE-defined TSSs and associated promoter activity data for all 3 subpopulations as well as putative enhancer regions for the classical and nonclassical subsets.
Digital gene expression, GO-term, and pathway analyses are largely in line with previous studies. In comparison with other transcriptome studies,13,14 intermediate subsets are most variable, which may reflect differences in FACS gating strategies used by individual groups. In addition to the previously noted upregulation of genes involved in antigen presentation, we also see a slight “activation” signature in the intermediate subset, including motif enrichment for a nuclear factor κB element. Because we are studying averages across cell populations, it is unclear, whether this is a general feature of intermediate monocytes or whether this signature derived from a small number of highly activated monocytes, which are absent or less frequent in both other populations. For similar reasons, our data can neither specifically support nor reject one of the 2 ontogeny models (linear differentiation vs independent progenitor) for classical and nonclassical monocytes.
The mapping of all TSSs as well as enhancer-associated histone modifications allowed us to perform comprehensive searches for subset-enriched sequence patterns. These analyses provide the first insights into motifs and potential binding factors that might be involved in regulating subset specific genes. The 2 major populations showed strikingly different motif signatures: classical monocytes were highly enriched for AP-1 and CEBP motifs that were previously also identified as key regulatory sequences in human and murine monocytes/macrophages,18,25 whereas the nonclassical signature included an enrichment of ETS motifs, a nuclear receptor motif resembling the consensus motif for NR4A1 (Nur77), which was recently described as a critical regulator of nonclassical monocytes in mice,10 as well as a number of general transcription factors that regulate housekeeping functions. The differential motif compositions as well as the CAGE-inferred transcription factor networks suggest that at least some of the subset-specific features are controlled either by expression changes of corresponding motif binding transcription factors or by differential signaling activities that are responsible for transcription factor activation.
Apart from providing a reference resource, the CAGE profiling identified biologically interesting differences between monocyte subsets, including the differential regulation of genes involved in energy metabolism. Although classical monocytes showed elevated levels of several genes involved in carbohydrate metabolism, including a major regulator of the glycolytic pathway (HIF1A), nonclassical monocytes demonstrated a higher transcriptional activity of many genes encoding components of the mitochondrial respiratory chain. Increased transcriptional activity of respiratory chain genes was not consistently associated with higher total messenger RNA levels (as measured by qRT-PCR) indicating significant posttranscriptional regulation of the corresponding transcripts. Nevertheless, nonclassical monocytes were also found to exhibit higher routine mitochondrial respiratory activity as compared with the classical subset. Although increased respiratory activity in nonclassical monocytes is accompanied by a simultaneous increase in CS activity, mitochondrial staining with a fluorescent dye revealed no difference in mitochondria content between subpopulations. Our findings are in line with a previous publication demonstrating an increased mitochondrial activity in CD16+ monocytes (including nonclassical and intermediate monocytes).50 The same study also noted a higher susceptibility toward apoptosis induced by reactive oxygen species, which correlated with lower expression of glutathione (GSH)-metabolizing genes such as GSH peroxidase (GSP1) and microsomal GSH S-transferase (MGST1),50 2 oxidative damage-limiting genes that also show weaker CAGE signals in nonclassical monocytes in the present study.
Although it is likely that the purification procedure constitutes a significant stress factor that might influence cell metabolism and general fitness of the purified cell populations, conditions were kept similar for all subpopulations, and the observed differences likely represent relevant features of monocyte subpopulations. In fact, a bias or priming of classical monocytes toward carbohydrate-based energy metabolism could be directly related to their subset-specific functions. It is known that classical monocytes rapidly extravasate during inflammation, a property largely attributed to high expression of the chemokine receptor CCR2.51 An immediate and energy-consuming response to many bacteria involves the production of reactive oxygen species via the phagosomal NADPH-oxidase–dependent respiratory burst, which was one of the first subset-specific features identified in classical monocytes (reviewed in Grage-Griebenow et al52 ). However, at sites of inflammation, monocytes encounter low oxygen levels, and it may be necessary to use carbohydrate metabolism (eg, to generate NADPH through the pentose phosphate pathway to fuel the respiratory burst). In contrast, nonclassical monocytes were previously shown to “patrol” blood vessels and specifically respond to viral infections.11 Whether the metabolic bias of nonclassical monocytes toward higher respiratory activity is related to these or other specific functions of this subset is currently unclear. Further studies will be required to clarify the physiological role of this phenomenon.
In conclusion, this study provides comprehensive insights into monocyte regulomes. The data are easily accessible through the UCSC Genome Browser and will provide a reference resource for monocyte biologists. In addition, our data indicate striking metabolomic differences between monocyte subpopulations that are likely linked with subset-specific functions.
This article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Johanna Raithel for excellent technical assistance, Christine Wells (UQ and Griffith University, Brisbane, Australia) for critically reading the manuscript and fruitful discussions, all members of the FANTOM5 consortium for contributing to the generation of samples and analysis of the data set, and GeNAS for data production.
FANTOM5 was made possible by a research grant for RIKEN Omics Science Center from MEXT (Y.H.) and an Innovative Cell Biology by Innovative Technology (Cell Innovation Program) grant from MEXT, Japan (Y.H.). This work was also supported by a research grant from MEXT to RIKEN Center for Life Science Technologies and from MEXT to RIKEN Preventive Medicine and Diagnosis Innovation Program. This project was funded by grants from the Deutsche Forschungsgemeinschaft (DFG Re1310/9, /13) (M.R.), BayImmuNet (M.E.), the Jose Carreras Foundation (FACS-Sorting Facility), and the Rudolf Bartling Foundation (M.R.).
Authorship
Contribution: C.S. performed experiments and computational analyses and wrote parts of the manuscript; K.R. performed experiments and contributed to manuscript writing; R.E. isolated the cells; K.P. performed experiments; P.H., R.A., M.P.K., and M.E. contributed to planning and supervision; P.J.B. performed computational analyses; M.I. and S.N.-S. were responsible for CAGE data production; T.L. was responsible for tag mapping and performed network analyses; H.K. managed the data handling; P.C., H.S., Y.H., and A.R.R.F. were responsible for FANTOM5 management and concept; D.A.H. contributed to data interpretation and manuscript writing; and M.R. initiated, planned, and supervised the study, performed computational analyses, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for Christian Schmidl is Research Center for Molecular Medicine of the Austrian Academy of Sciences (CeMM), 1090 Vienna, Austria.
Correspondence: Michael Rehli, Department of Internal Medicine III, University Hospital Regensburg, D-93042 Regensburg, Germany; e-mail: michael.rehli@ukr.de.
References
Author notes
*RIKEN Omics Science Center ceased to exist as of April 1, 2013 due to RIKEN reorganization.

![Figure 3. Monocyte subset-specific active enhancer signatures. (A) Bubble-plot representation of CAGE-TSS activity around subset-specific enhancer candidate regions showing at least 3-fold different H3K27ac signals. The bubble plots encode 3 quantitative parameters per CAGE cluster: distance from the putative enhancer, log10 of fold change in CAGE cluster tag count between classical and nonclassical monocytes (y-axis), and the absolute CAGE cluster tag count of the monocyte subset with the highest expression level (bubble diameter). There is a clear bias for the putative enhancer elements to associate with CAGE clusters upregulated in the corresponding cell type (P < .001, Wilcoxon signed-rank test). (B) Putative subset-specific enhancer region of the human CD14 locus. On top, UCSC Genome Browser tracks covering the entire CD14 locus are shown including epigenetic data (H3K4me1 and H3K27ac), positions of transcription factor binding sites for PU.1 and C/EBPβ in total monocytes,18 positions of bidirectional enhancers identified from the FANTOM5 expression atlas,44 HeliScopeCAGE data (monocyte subsets are indicated by coloring: classical, red; intermediate, green; nonclassical, blue), GencodeV10 gene annotation, 5′-RACE–based annotation of a novel TMOC6 transcript, and positions of genomic intervals that were used for enhancer reporter assays. The bottom panel shows relative luciferase activities (enhancer construct/CD14 promoter only [control]) of individual enhancer constructs in a monocytic (THP-1) and a T-cell (Jurkat) cell line. A myeloid-specific set of CD14 enhancers was detected between regions 6 and 8.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/123/17/10.1182_blood-2013-02-484188/4/m_e90f3.jpeg?Expires=1765885161&Signature=TK~vJA6-oUMsHj4spWTMoIkV8r2oTtoMXg~PZCCNeAFAOvusoulpvr8VWEJY6e8U0rAXTq3HZFYbIrsjegrWDTQwETjoJ5AJGDeYaroW9ppF6jaGoLdnhj2-0OBKRkIMU-8EczrVeVoekZ0f2EQIFNFihbcHPwqCMt9JLmnnpYr6gAmMbMk3mVPali7jV-ZIdVRF9igkvOVBiM23YP6Q~WnjF9LJAcFXmEEf6Rp0qGrTkIZiDAsEkk60SSkMUw8TXSFRyJ8FZQzSekUq8gqnxpohI-HT03nJCjnSgeRTyNrvJQOq5Vmq2SdQZlby7LVq2irfboUNXpZUg4dQ7Tr8rw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)