Key Points
Alternatively activated macrophages derived from monocytes and tissue macrophages have distinct transcriptional profiles and phenotypes.
Monocyte-derived AAMs are more involved with immune regulation than tissue-derived AAMs.
Abstract
Macrophages adopt an alternatively activated phenotype (AAMs) when activated by the interleukin-4receptor(R)α. AAMs can be derived either from proliferation of tissue resident macrophages or recruited inflammatory monocytes, but it is not known whether these different sources generate AAMs that are phenotypically and functionally distinct. By transcriptional profiling analysis, we show here that, although both monocyte and tissue-derived AAMs expressed high levels of Arg1, Chi3l3, and Retnla, only monocyte-derived AAMs up-regulated Raldh2 and PD-L2. Monocyte-derived AAMs were also CX3CR1-green fluorescent protein (GFP)high and expressed CD206, whereas tissue-derived AAMs were CX3CR1-GFP and CD206 negative. Monocyte-derived AAMs had high levels of aldehyde dehydrogenase activity and promoted the differentiation of FoxP3+ cells from naïve CD4+ cells via production of retinoic acid. In contrast, tissue-derived AAMs expressed high levels of uncoupling protein 1. Hence monocyte-derived AAM have properties associated with immune regulation, and the different physiological properties associated with AAM function may depend on the distinct lineage of these cells.
Introduction
There has recently been a major paradigm shift in the field of macrophage biology with the recognition that tissue resident macrophages can be established independently of definitive hematopoiesis.1-4 These cells are of embryonic origin and are maintained throughout life by proliferative self-renewal.5 In contrast, macrophages infiltrating the tissues during an inflammatory response are derived from blood monocytes.6,7 This paradigm holds true in many tissues including the serous cavities1,3 but not for the intestines8,9 and the skin,10 where the resident cells are of bone marrow origin.
During T helper 2 (TH2)-mediated immune responses, interleukin (IL)-4 and/or IL-13 can induce macrophage proliferation, leading to expansion beyond steady-state levels.11,12 Signaling through the IL-4receptor(R)α also leads to a state of alternative activation.13,14 Alternatively activated macrophages (AAMs) are a critical component of type 2 immunity during helminth infection15 and allergic responses.16 However, type 2 immune responses extend beyond just infection and autoimmunity17 and contribute to the maintenance of tissue homeostasis,16 damage repair,18 and metabolic homeostasis.19
IL-4 will induce proliferation and alternative activation of macrophages regardless of their embryonic or bone marrow origins.11 This raises critical questions about the contribution of tissue-resident macrophages vs blood-derived macrophages to the diverse processes associated with AAMs. Bone marrow chimeras in which the peritoneal and pleural cavity cells are shielded from irradiation, and thus remain of host origin, allow us to distinguish cells of bone marrow vs resident origins.11 Using this method, we have demonstrated that IL-4 induces the proliferation and expansion of resident peritoneal macrophages, whereas delivery of IL-4 plus thioglycollate caused recruitment and proliferation of blood monocyte-derived macrophages.11 Here, we demonstrate that AAMs derived from proliferation of local tissue resident macrophages are phenotypically and functionally distinct from AAMs derived through recruitment of inflammatory monocytes.
Materials and methods
Mice treatment and infections
C57BL/6, Stat6−/− (Jackson Laboratories) and CX3CR1GFP/+ mice (kindly provided by Dr Dan Littman) were treated with IL-4/anti-IL-4 monoclonal antibody (mAb) complexes (IL-4c), prepared as described previously.11 Mice were injected intraperitoneally (i.p.) with IL-4c on days 0 and 2. Mice were also treated with 4% thioglycollate alone or in combination with IL-4c for coadministration experiments. Mice were euthanized on day 4. Mice were infected subcutaneously with 25 Litomososides sigmodontis L3,20 and analysis of pleural cavity cells was performed 12 days after infection. For Schisotosoma mansoni infection, mice were infected percutaneously on the abdominal surface with 25 to 35 cercariae of a Puerto Rican strain (Biomedical Research Institute, Rockville, MD), and analysis of liver leukocytes was performed 8 weeks after infection. This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals. All animal procedures were approved by the New York University Institutional Animal Care and Use Committee under protocol numbers 131004 and 130504, as well as in accordance with the United Kingdom Home Office requirements.
Flow cytometry
Cells were stained with LIVE/DEAD Aqua or Blue (Invitrogen), blocked with 4 μg/mL aCD16/32 (2.4G2; Bioxcell), and stained with CD11b eFluor450 (eBioscience), MHCII APC-Cy7 (Biolegend), PD-L2 PE (BD Bioscience, Biolegend), F4/80 PE-Cy7 (eBioscience), CD206 APC (Biolegend), and Siglec-F, DX5, B220, and CD3 PE (BD Bioscience). Cells were acquired on an LSR II (BD Biosciences) and analyzed using FlowJo software (Treestar). For 5-ethynyl-2′-deoxyuridine (EdU) labeling, mice were injected i.p. with 0.5 mg EdU (Invitrogen) 3 hours prior to euthanasia and stained for EdU according to the manufacturer’s instructions. Aldehyde dehydrogenase (ALDH) activity was measured using the ALDEFLUOR staining kit (StemCell Technologies) with or without the ALDH inhibitor, diethylamino-benzaldehyde (DEAB) (at a final concentration of 15 µM) as control.
Foxp3+ T-regulatory cell differentiation assay
4 × 105 naïve T cells isolated from lymph nodes using the Naive CD4+ T Cell Isolation Kit II (Miltenyi Biotec) were cultured together with 2 × 105 peritoneal macrophages and 1 µg/mL of soluble anti-CD3 and 5 ng/mL recombinant human IL-2 (R&D) in complete RPMI. On day 3, cultures were supplemented with fresh medium containing 5 ng/mL IL-2. Retinoic acid (RA) receptor inhibitor LE540 (Wako Chemicals USA) was added to some culture wells. To track cell division, T cells were labeled with Violet CellTracker (Invitrogen). On day 6, cells were stained for Foxp3+ cells by intracellular nuclear staining.
Microarray analysis of sorted cells
CD11b and F4/80 double positive cells (supplemental Figure 3, available on the Blood Web site) sorted using a BD FACSAria cell were analyzed by using Sureprint G3 Mouse GE 8x60K array (Agilent) in one color (Cy3)-based gene expression analysis (Agilent). Processing and downstream analysis of microarray data (Gene Expression Omnibus accession number GSE54679) was performed with R/Bioconductor and associated packages and SAM:Significance Analysis of Microarrays.21 Background correction to log-transformed expression values was performed, and the expression values of each array was scaled so that the median absolute values are equal for all arrays. The Clustering of Large Applications function from R was used to find the best clustering solution that coincided with the global minimum Bayesian information critera score and a locally maximum silhouette width. Each cluster was assessed by measuring the Gene Ontology (GO) term over-representation by the hypergeometric distribution, as implemented in the GOstats package using gene annotations from the mouse.db0 package.
RT-PCR and western blot
Quantitative reverse transcription-polymerase chain reaction (RT-PCR) was performed using the SYBR Green qPCR kit (Applied Biosystems) and normalized to the housekeeping gene Gapdh by comparative Ct. For western blots, anti-UCP1 antibody (ab-10983; Abcam) and antiglyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody (sc-32233; SCBT) were used.
Statistical analysis
Significance between groups was determined by analysis of variance (ANOVA) plus Bonferroni or Dunnett’s correction for multiple comparisons.
Results
Peritoneal AAMs derived from inflammatory monocytes and tissue macrophages both proliferate and express markers of alternative activation
Using shielded bone marrow chimeras, we previously showed that injection of IL-4c into the pleural cavity leads to the expansion of the resident cell population independently of the bone marrow, whereas simultaneous injection of IL-4c with thioglycollate (Thio+IL-4c) generates a large population of AAMs that are derived from inflammatory blood monocytes.11 We applied this system to the peritoneal cavity to compare AAMs derived from proliferating tissue resident macrophages (IL-4c) with AAMs derived from inflammatory monocytes (Thio+IL-4c). Resident macrophages from naïve untreated animals (Res) and macrophages recruited by thioglycollate alone (Thio) were used as controls. We confirmed by 3-hour pulse labeling and EdU staining that both IL-4c treatment and Thio+IL-4c treatment can induce macrophage proliferation, although Thio+IL-4c induced macrophages proliferated to a lesser degree with lower proportion of cells in S phase (Figure 1A-B). Less proliferation may be due to reduced availability of IL-4 per cell, as the recruited population will quickly outnumber the resident cells in the Thio+IL-4c-treated mice.
IL-4c and Thio+IL-4c induces alternatively activated macrophages with distinct phenotypes. Mice were untreated (naïve/resident) or injected i.p. with IL-4c alone (IL-4c) or thioglycollate alone (Thio) or Thio and IL-4c (Thio+IL-4c) on day 0 and then with IL-4c again (for IL-4c and Thio+IL-4c) on day 2, and PEC was analyzed on day 4. (A) Representative FACS plots shows EdU incorporation by peritoneal macrophages after treatment. EdU was injected 3 hours before analysis. (B) Quantification of EdU incorporation in macrophages. Results are representative of 3 independent experiments. (C) RNA of peritoneal macrophages was isolated for RT-PCR analysis for expression of Chi3l3, Arg1, Retnla, and Raldh2 and normalized to expression of GAPDH. Graphs depict mean ± standard error of the mean of individual mice pooled from 5 to 6 independent experiments. (D) EdU incorporation by peritoneal macrophages after injection of wild-type Stat6+/+ mice or Stat6−/− mice with IL-4c alone, Thio, or Thio+IL-4c. (E) Quantification of EdU incorporation in macrophages. (F) Total number of F4/80+ macrophages recovered. Results shown are representative of 2 separate experiments. (G) Representative FACS plots gated on CD11b+ cells for CX3CR1GFP/+ reporter mice treated as above. (H) The median fluorescent intensity (MFI) of GFP, gated on CD11b+ cells from the peritoneal cavity of individual mice. Data are representative of 3 independent experiments. *P < .05 and **P < .01 as determined by ANOVA.
IL-4c and Thio+IL-4c induces alternatively activated macrophages with distinct phenotypes. Mice were untreated (naïve/resident) or injected i.p. with IL-4c alone (IL-4c) or thioglycollate alone (Thio) or Thio and IL-4c (Thio+IL-4c) on day 0 and then with IL-4c again (for IL-4c and Thio+IL-4c) on day 2, and PEC was analyzed on day 4. (A) Representative FACS plots shows EdU incorporation by peritoneal macrophages after treatment. EdU was injected 3 hours before analysis. (B) Quantification of EdU incorporation in macrophages. Results are representative of 3 independent experiments. (C) RNA of peritoneal macrophages was isolated for RT-PCR analysis for expression of Chi3l3, Arg1, Retnla, and Raldh2 and normalized to expression of GAPDH. Graphs depict mean ± standard error of the mean of individual mice pooled from 5 to 6 independent experiments. (D) EdU incorporation by peritoneal macrophages after injection of wild-type Stat6+/+ mice or Stat6−/− mice with IL-4c alone, Thio, or Thio+IL-4c. (E) Quantification of EdU incorporation in macrophages. (F) Total number of F4/80+ macrophages recovered. Results shown are representative of 2 separate experiments. (G) Representative FACS plots gated on CD11b+ cells for CX3CR1GFP/+ reporter mice treated as above. (H) The median fluorescent intensity (MFI) of GFP, gated on CD11b+ cells from the peritoneal cavity of individual mice. Data are representative of 3 independent experiments. *P < .05 and **P < .01 as determined by ANOVA.
We next examined expression of the well-defined markers of alternative activation, arginase I (Arg1), resistin-like molecule α (Retnla), and Ym1 (or Chi3l3), in both types of AAMs. By real-time PCR analysis, expression of all 3 genes was highly up-regulated in both IL-4c- and Thio+IL-4c-induced macrophages (Figure 1C). This was consistent with our previous studies in the pleural cavity in which both IL-4c and Thio+IL-4c led to extensive alternative activation as measured by high percentages of resistin-like molecule α (RELMα)+ and Ym1+ macrophages.11 Stat6 is essential for alternative activation,22,23 but its role in macrophage proliferation has not been previously demonstrated. Both Thio+IL-4c and IL-4c activated macrophages from Stat6−/− mice failed to increase proliferation and incorporate EdU in contrast to wild-type littermates (Figure 1D-E), confirming Stat6 dependence. The failure to proliferate was reflected in reduced total cell numbers especially among F4/80+ macrophages (Figure 1F).
In CX3CR1-green fluorescent protein (GFP)/+ reporter mice, CX3CR1-GFP is highly expressed by blood monocytes but not by tissue resident peritoneal macrophages.24 To validate the different origins of cells derived from IL-4c vs Thio+IL-4c, we injected CX3CR1-GFP/+ reporter mice with these reagents. Gating on the CD11b+ cells, the majority of macrophages induced by IL-4c alone were F480high and CX3CR1-GFPlow, similar to the resident population in naïve mice. In contrast, the AAMs induced by Thio+IL-4c were F480int and CX3CR1-GFPhigh (Figure 1G-H). These results are consistent with a monocyte origin for AAMs induced by Thio+IL-4c (F480int), whereas those induced by IL-4c alone are derived from resident macrophages (F480high). Notably, there is a small CX3CR1-GFPhigh F4/80int macrophage population in naïve mice and IL-4c alone-treated mice that is likely of monocyte origin.25
We confirmed that IL-4c induces expansion of the resident cell population in the peritoneal cavity independently of the bone marrow by generating bone marrow chimeras in which the serous cavities are shielded from irradiation and assessing the chimerism in the blood vs the peritoneal cavity (supplemental Figure 1). The blood chimerism was ∼30% of donor origin, whereas only 1% to 2% of macrophages in the peritoneal cavity were of donor origin in both phosphate-buffered saline (PBS) and IL-4c-injected mice. Similarly, by intravenous transfer of purified Ly6CHigh monocytes from CX3CR1-GFP/+ mice (CD45.2) into congenic recipient CD45.1 hosts, we confirmed that Thio+IL-4c but not IL-4c alone induces monocyte recruitment and differentiation (supplemental Figure 1).
AAMs derived from monocytes and tissue macrophages are transcriptionally distinct
We next decided to generate a detailed transcriptional profile of peritoneal AAMs of distinct origins. We fluorescence-activated cell sorter (FACS) sorted pure populations of CD11b+, F4/80High cells for resident peritoneal macrophages and IL-4c-induced macrophages and CD11b+, F4/80Int cells for Thio- and Thio+IL-4c-induced macrophages (supplemental Figure 2). These 4 populations were subjected to gene expression profiling analysis with whole genome microarrays (Figure 2). Unsupervised hierarchical clustering analysis showed coclustering of IL-4c-induced F4/80High AAMs with resident F4/80High macrophages from untreated mice (Figure 2A), whereas AAMs induced by Thio+IL-4c coclustered with Thio induced macrophages (Figure 2A). When AAMs induced by IL-4c were directly compared with Thio+IL-4c-induced AAMs, expression across all probes was remarkably different between these 2 types of macrophages (Figure 2B), which is consistent with their different cellular lineages. A detailed analysis of some genes previously described in AAMs (Figure 2C) confirmed the up-regulation of arginase I, RELMα, and Ym1 (or Chi3l3), as well as Tgm226 and Klf427 in both types of AAMs (Figure 2C). Tgm2 was recently identified as a universal AAM marker for both humans and mice.26 Aldh1a2 (Raldh2), pdcd1lg2 (PDL2), Socs2, IL-31ra, Ccl17, Ch25h, Jag2, and Ccl22 were more highly expressed on Thio+IL-4c-induced AAM (Figure 2C).
Gene expression profiling of monocyte-derived and tissue macrophage-derived AAMs. (A) Unsupervised hierarchical clustering of transcriptional profiles, displayed as a heat map of log-transformed expression values, from FACS-purified (CD11b+F4/80+) macrophages of mice untreated (Res) or treated with IL-4c alone (IL-4c) or thioglycollate alone (Thio) or Thio and IL-4c (Thio+IL-4c). (B) Comparison of probe expression for AAMs induced by IL-4c alone or Thio+IL-4c, indicating that most genes are differentially expressed. (C) Heat map of log-transformed expression values for a subset of genes associated with AAMs. (D) Venn diagrams showing the overlap of genes up-regulated and down-regulated by IL-4 in monocyte-derived (Thio+IL-4c vs Thio) compared with tissue-derived AAMs (IL-4c vs resident). List of genes are shown in Tables 1 to 8.
Gene expression profiling of monocyte-derived and tissue macrophage-derived AAMs. (A) Unsupervised hierarchical clustering of transcriptional profiles, displayed as a heat map of log-transformed expression values, from FACS-purified (CD11b+F4/80+) macrophages of mice untreated (Res) or treated with IL-4c alone (IL-4c) or thioglycollate alone (Thio) or Thio and IL-4c (Thio+IL-4c). (B) Comparison of probe expression for AAMs induced by IL-4c alone or Thio+IL-4c, indicating that most genes are differentially expressed. (C) Heat map of log-transformed expression values for a subset of genes associated with AAMs. (D) Venn diagrams showing the overlap of genes up-regulated and down-regulated by IL-4 in monocyte-derived (Thio+IL-4c vs Thio) compared with tissue-derived AAMs (IL-4c vs resident). List of genes are shown in Tables 1 to 8.
We then performed supervised comparisons by statistical analyses of microarrays of IL-4c-induced AAMs vs resident macrophages and Thio+IL-4c-induced AAMs vs Thio-induced macrophages to identify significantly different genes with a false discovery rate of 0%. With this cutoff, 758 genes were up-regulated in IL-4c-treated macrophages (Table 1) compared with controls, and 368 genes were up-regulated in Thio+IL-4c-treated macrophages (Table 2) compared with Thio-induced macrophages. One hundred fifty-three genes were shared between these 2 groups (Table 3; Figure 2D). Genes up-regulated in the IL-4c-treated macrophages were enriched by GO analysis for the biological processes involved in cellular replication (cell cycle, mitosis, DNA replication, etc) (Figure 3A). Consistent with increased proliferation in both IL-4c and Thio+IL-4c activated macrophages, genes involved in cellular replication and metabolism were up-regulated in both types of AAMs (Figure 3C). Many of the genes up-regulated in Thio+IL-4c-treated macrophages were unclassified in terms of their biological process (Figure 3B). In contrast to IL-4c-induced macrophages, up-regulated genes classified as part of the immune system were enriched in Thio+IL-4c-induced macrophages. Molecular function GO analysis revealed that cytokine (IL-6, Cish, IL31Ra, Socs6, Socs2) and chemokine (Ccl17, Ccl2, Ccl7, ccl12, and Ccl24) activity were the main functions up-regulated in Thio+IL-4c AAMs (Figure 3D), but these functions were not up-regulated in IL-4c-treated macrophages; instead, chemokine (Ccl3, Cxcl1, Cxcl2, and Cxcl13) activity was down-regulated (supplemental Figure 3; Tables 4-6).
List of top 50 genes up-regulated by IL-4 in resident macrophages
| Gene symbol . | Description . | Fold change . |
|---|---|---|
| AA467197 | Expressed sequence AA467197 | 665.9877 |
| Chi3l4 | Chitinase-like 4 or Ym2 | 526.5159 |
| Aqp3 | Aquaporin 3 | 295.7499 |
| Sprr2a2 | Small proline-rich protein 2A2 | 149.7646 |
| Cst7 | Cystatin F (leukocystatin) | 135.3837 |
| Havcr1 | Hepatitis A virus cellular receptor 1 | 116.3776 |
| Mall | Mal, T cell differentiation protein-like | 109.5096 |
| Ddx4 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 | 107.4824 |
| Tmigd1 | Transmembrane and immunoglobulin domain containing 1 | 92.0161 |
| Timd2 | T-cell immunoglobulin and mucin domain containing 2 | 90.5118 |
| Chi3l3 | Chitinase-like 3 or Ym1 | 81.1482 |
| Ucp1 | Uncoupling protein 1 (mitochondrial, proton carrier) | 74.1526 |
| Serpinb7 | Serine (or cysteine) peptidase inhibitor, clade B, member 7 | 72.4033 |
| Tcf23 | Transcription factor 23 | 70.0783 |
| Ear10 | Eosinophil-associated, ribonuclease A family, member 10 | 66.9156 |
| Ear11 | Eosinophil-associated, ribonuclease A family, member 11 | 63.0005 |
| Gm4610 | Predicted gene 4610 | 53.6569 |
| Syt10 | Synaptotagmin X | 52.9130 |
| Gm5150 | Predicted gene 5150 | 49.2676 |
| Klk1b11 | Kallikrein 1-related peptidase b11 | 47.2572 |
| Car12 | Carbonic anyhydrase 12 | 43.5869 |
| Gatm | Glycine amidinotransferase (l-arginine:glycine amidinotransferase) | 42.6830 |
| I830127L07Rik | RIKEN cDNA I830127L07 gene | 38.1699 |
| LOC101056540 | Unknown | 35.2395 |
| Asns | Asparagine synthetase | 33.8962 |
| Rrm2 | Ribonucleotide reductase M2 | 32.8288 |
| Chchd10 | Coiled-coil-helix-coiled-coil-helix domain containing 10 | 31.9263 |
| Flt1 | FMS-like tyrosine kinase 1 | 31.6849 |
| Slc7a2 | Solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 | 30.7286 |
| Galnt6 | UDP-N-acetyl-α-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 | 30.3134 |
| Trib3 | Tribbles homolog 3 (Drosophila) | 27.4894 |
| A530064D06Rik | RIKEN cDNA A530064D06 gene | 26.7710 |
| Ccrn4l | CCR4 carbon catabolite repression 4-like (S cerevisiae) | 25.7457 |
| Grhl3 | Grainyhead-like 3 (Drosophila) | 25.1532 |
| Entpd3 | Ectonucleoside triphosphate diphosphohydrolase 3 | 24.6177 |
| Kcnn4 | Potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 | 22.9964 |
| Fxyd6 | FXYD domain-containing ion transport regulator 6 | 22.4216 |
| Hist1h1b | Histone cluster 1, H1b | 22.4066 |
| Arg1 | Arginase, liver | 22.1165 |
| Oasl1 | 2'-5′ oligoadenylate synthetase-like 1 | 21.4259 |
| Gfra2 | Glial cell line derived neurotrophic factor family receptor α 2 | 20.4561 |
| Anpep | Alanyl (membrane) aminopeptidase | 20.3326 |
| Shcbp1 | Shc SH2-domain binding protein 1 | 19.8717 |
| Sema3b | Sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B | 19.7311 |
| Pbk | PDZ binding kinase | 19.4258 |
| Spp1 | Secreted phosphoprotein 1 | 19.0967 |
| Ccna2 | Cyclin A2 | 18.2413 |
| Nuf2 | NUF2, NDC80 kinetochore complex component, homolog (S cerevisiae) | 18.0971 |
| Ccnf | Cyclin F | 17.6265 |
| Plxna2 | Plexin A2 | 17.4434 |
| Gene symbol . | Description . | Fold change . |
|---|---|---|
| AA467197 | Expressed sequence AA467197 | 665.9877 |
| Chi3l4 | Chitinase-like 4 or Ym2 | 526.5159 |
| Aqp3 | Aquaporin 3 | 295.7499 |
| Sprr2a2 | Small proline-rich protein 2A2 | 149.7646 |
| Cst7 | Cystatin F (leukocystatin) | 135.3837 |
| Havcr1 | Hepatitis A virus cellular receptor 1 | 116.3776 |
| Mall | Mal, T cell differentiation protein-like | 109.5096 |
| Ddx4 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 | 107.4824 |
| Tmigd1 | Transmembrane and immunoglobulin domain containing 1 | 92.0161 |
| Timd2 | T-cell immunoglobulin and mucin domain containing 2 | 90.5118 |
| Chi3l3 | Chitinase-like 3 or Ym1 | 81.1482 |
| Ucp1 | Uncoupling protein 1 (mitochondrial, proton carrier) | 74.1526 |
| Serpinb7 | Serine (or cysteine) peptidase inhibitor, clade B, member 7 | 72.4033 |
| Tcf23 | Transcription factor 23 | 70.0783 |
| Ear10 | Eosinophil-associated, ribonuclease A family, member 10 | 66.9156 |
| Ear11 | Eosinophil-associated, ribonuclease A family, member 11 | 63.0005 |
| Gm4610 | Predicted gene 4610 | 53.6569 |
| Syt10 | Synaptotagmin X | 52.9130 |
| Gm5150 | Predicted gene 5150 | 49.2676 |
| Klk1b11 | Kallikrein 1-related peptidase b11 | 47.2572 |
| Car12 | Carbonic anyhydrase 12 | 43.5869 |
| Gatm | Glycine amidinotransferase (l-arginine:glycine amidinotransferase) | 42.6830 |
| I830127L07Rik | RIKEN cDNA I830127L07 gene | 38.1699 |
| LOC101056540 | Unknown | 35.2395 |
| Asns | Asparagine synthetase | 33.8962 |
| Rrm2 | Ribonucleotide reductase M2 | 32.8288 |
| Chchd10 | Coiled-coil-helix-coiled-coil-helix domain containing 10 | 31.9263 |
| Flt1 | FMS-like tyrosine kinase 1 | 31.6849 |
| Slc7a2 | Solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 | 30.7286 |
| Galnt6 | UDP-N-acetyl-α-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 | 30.3134 |
| Trib3 | Tribbles homolog 3 (Drosophila) | 27.4894 |
| A530064D06Rik | RIKEN cDNA A530064D06 gene | 26.7710 |
| Ccrn4l | CCR4 carbon catabolite repression 4-like (S cerevisiae) | 25.7457 |
| Grhl3 | Grainyhead-like 3 (Drosophila) | 25.1532 |
| Entpd3 | Ectonucleoside triphosphate diphosphohydrolase 3 | 24.6177 |
| Kcnn4 | Potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 | 22.9964 |
| Fxyd6 | FXYD domain-containing ion transport regulator 6 | 22.4216 |
| Hist1h1b | Histone cluster 1, H1b | 22.4066 |
| Arg1 | Arginase, liver | 22.1165 |
| Oasl1 | 2'-5′ oligoadenylate synthetase-like 1 | 21.4259 |
| Gfra2 | Glial cell line derived neurotrophic factor family receptor α 2 | 20.4561 |
| Anpep | Alanyl (membrane) aminopeptidase | 20.3326 |
| Shcbp1 | Shc SH2-domain binding protein 1 | 19.8717 |
| Sema3b | Sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B | 19.7311 |
| Pbk | PDZ binding kinase | 19.4258 |
| Spp1 | Secreted phosphoprotein 1 | 19.0967 |
| Ccna2 | Cyclin A2 | 18.2413 |
| Nuf2 | NUF2, NDC80 kinetochore complex component, homolog (S cerevisiae) | 18.0971 |
| Ccnf | Cyclin F | 17.6265 |
| Plxna2 | Plexin A2 | 17.4434 |
List of top 50 genes up-regulated by IL-4 in monocyte-derived macrophages
| Gene symbol . | Description . | Fold change . |
|---|---|---|
| Ccl24 | Chemokine (C-C motif) ligand 24 | 308.1040 |
| Ddx4 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 | 147.2503 |
| Ear11 | Eosinophil-associated, ribonuclease A family, member 11 | 129.8195 |
| Tuba8 | Tubulin, α 8 | 114.0360 |
| Ear10 | Eosinophil-associated, ribonuclease A family, member 10 | 109.5308 |
| Chi3l4 | Chitinase-like 4 or Ym2 | 95.2618 |
| Ear1 | Eosinophil-associated, ribonuclease A family, member 1 | 90.4369 |
| Ear12 | Eosinophil-associated, ribonuclease A family, member 12 | 88.9709 |
| Cdh1 | Cadherin 1 | 73.2850 |
| Il4i1 | Interleukin 4 induced 1 | 71.5692 |
| AA467197 | Expressed sequence AA467197 | 68.8517 |
| Asns | Asparagine synthetase | 60.0067 |
| Ch25h | Cholesterol 25-hydroxylase | 59.7103 |
| Cish | Cytokine inducible SH2-containing protein | 59.3660 |
| Gm6756 | Predicted gene 6756 | 53.0013 |
| Ear2 | Eosinophil-associated, ribonuclease A family, member 2 | 47.4012 |
| Vdr | Vitamin D receptor | 42.3885 |
| Timp1 | Tissue inhibitor of metalloproteinase 1 | 40.8995 |
| Pi16 | Peptidase inhibitor 16 | 39.9049 |
| Fbp1 | Fructose bisphosphatase 1 | 37.6432 |
| Cst7 | Cystatin F (leukocystatin) | 36.9475 |
| Klk8 | Kallikrein related-peptidase 8 | 36.9350 |
| Gfra2 | Glial cell line derived neurotrophic factor family receptor α 2 | 36.3341 |
| Phgdh | 3-phosphoglycerate dehydrogenase | 36.1081 |
| Tarm1 | T cell-interacting, activating receptor on myeloid cells 1 | 36.1055 |
| Ramp3 | Receptor (calcitonin) activity modifying protein 3 | 31.2576 |
| A530064D06Rik | RIKEN cDNA A530064D06 gene | 26.3184 |
| Slc7a2 | Solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 | 24.4996 |
| Msx3 | msh homeobox 3 | 21.9209 |
| Nlrp1a | NLR family, pyrin domain containing 1A | 21.6752 |
| Ccl7 | Chemokine (C-C motif) ligand 7 | 21.4119 |
| Car6 | Carbonic anhydrase 6 | 20.7420 |
| Cebpe | CCAAT/enhancer binding protein (C/EBP), ε | 20.2090 |
| Aldh1a2 | Aldehyde dehydrogenase family 1, subfamily A2 | 19.2205 |
| Ccl17 | Chemokine (C-C motif) ligand 17 | 19.2074 |
| Mthfd2 | Methylenetetrahydrofolate dehydrogenase (NAD+ dependent), methenyltetrahydrofolate cyclohydrolase | 16.9681 |
| Aqp3 | Aquaporin 3 | 16.4622 |
| Alox15 | Arachidonate 15-lipoxygenase | 15.4346 |
| Retnla | Resistin like α | 15.3394 |
| Ltb4r1 | Leukotriene B4 receptor 1 | 15.2231 |
| Retnlg | Resistin like γ | 15.1644 |
| Flt1 | FMS-like tyrosine kinase 1 | 14.4730 |
| Olr1 | Oxidized low density lipoprotein (lectin-like) receptor 1 | 14.4345 |
| Cd81 | CD81 antigen | 14.2763 |
| Slc7a5 | Solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 | 13.8395 |
| Fcrls | Fc receptor-like S, scavenger receptor | 12.9906 |
| Ap4e1 | Adaptor-related protein complex AP-4, ε 1 | 12.5573 |
| Hic1 | Hypermethylated in cancer 1 | 12.2985 |
| Rnase6 | Ribonuclease, RNase A family, 6 | 12.1713 |
| Slc30a4 | Solute carrier family 30 (zinc transporter), member 4 | 12.0286 |
| Gene symbol . | Description . | Fold change . |
|---|---|---|
| Ccl24 | Chemokine (C-C motif) ligand 24 | 308.1040 |
| Ddx4 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 | 147.2503 |
| Ear11 | Eosinophil-associated, ribonuclease A family, member 11 | 129.8195 |
| Tuba8 | Tubulin, α 8 | 114.0360 |
| Ear10 | Eosinophil-associated, ribonuclease A family, member 10 | 109.5308 |
| Chi3l4 | Chitinase-like 4 or Ym2 | 95.2618 |
| Ear1 | Eosinophil-associated, ribonuclease A family, member 1 | 90.4369 |
| Ear12 | Eosinophil-associated, ribonuclease A family, member 12 | 88.9709 |
| Cdh1 | Cadherin 1 | 73.2850 |
| Il4i1 | Interleukin 4 induced 1 | 71.5692 |
| AA467197 | Expressed sequence AA467197 | 68.8517 |
| Asns | Asparagine synthetase | 60.0067 |
| Ch25h | Cholesterol 25-hydroxylase | 59.7103 |
| Cish | Cytokine inducible SH2-containing protein | 59.3660 |
| Gm6756 | Predicted gene 6756 | 53.0013 |
| Ear2 | Eosinophil-associated, ribonuclease A family, member 2 | 47.4012 |
| Vdr | Vitamin D receptor | 42.3885 |
| Timp1 | Tissue inhibitor of metalloproteinase 1 | 40.8995 |
| Pi16 | Peptidase inhibitor 16 | 39.9049 |
| Fbp1 | Fructose bisphosphatase 1 | 37.6432 |
| Cst7 | Cystatin F (leukocystatin) | 36.9475 |
| Klk8 | Kallikrein related-peptidase 8 | 36.9350 |
| Gfra2 | Glial cell line derived neurotrophic factor family receptor α 2 | 36.3341 |
| Phgdh | 3-phosphoglycerate dehydrogenase | 36.1081 |
| Tarm1 | T cell-interacting, activating receptor on myeloid cells 1 | 36.1055 |
| Ramp3 | Receptor (calcitonin) activity modifying protein 3 | 31.2576 |
| A530064D06Rik | RIKEN cDNA A530064D06 gene | 26.3184 |
| Slc7a2 | Solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 | 24.4996 |
| Msx3 | msh homeobox 3 | 21.9209 |
| Nlrp1a | NLR family, pyrin domain containing 1A | 21.6752 |
| Ccl7 | Chemokine (C-C motif) ligand 7 | 21.4119 |
| Car6 | Carbonic anhydrase 6 | 20.7420 |
| Cebpe | CCAAT/enhancer binding protein (C/EBP), ε | 20.2090 |
| Aldh1a2 | Aldehyde dehydrogenase family 1, subfamily A2 | 19.2205 |
| Ccl17 | Chemokine (C-C motif) ligand 17 | 19.2074 |
| Mthfd2 | Methylenetetrahydrofolate dehydrogenase (NAD+ dependent), methenyltetrahydrofolate cyclohydrolase | 16.9681 |
| Aqp3 | Aquaporin 3 | 16.4622 |
| Alox15 | Arachidonate 15-lipoxygenase | 15.4346 |
| Retnla | Resistin like α | 15.3394 |
| Ltb4r1 | Leukotriene B4 receptor 1 | 15.2231 |
| Retnlg | Resistin like γ | 15.1644 |
| Flt1 | FMS-like tyrosine kinase 1 | 14.4730 |
| Olr1 | Oxidized low density lipoprotein (lectin-like) receptor 1 | 14.4345 |
| Cd81 | CD81 antigen | 14.2763 |
| Slc7a5 | Solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 | 13.8395 |
| Fcrls | Fc receptor-like S, scavenger receptor | 12.9906 |
| Ap4e1 | Adaptor-related protein complex AP-4, ε 1 | 12.5573 |
| Hic1 | Hypermethylated in cancer 1 | 12.2985 |
| Rnase6 | Ribonuclease, RNase A family, 6 | 12.1713 |
| Slc30a4 | Solute carrier family 30 (zinc transporter), member 4 | 12.0286 |
List of 50 genes regulated by IL-4 in both resident and monocyte-derived macrophages
| Gene symbol . | Description . | Fold change* . |
|---|---|---|
| AA467197 | Expressed sequence AA467197 | 367.4197 |
| Chi3l4 | Chitinase-like 4 or Ym2 | 310.8889 |
| Aqp3 | Aquaporin 3 | 156.1061 |
| Ddx4 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 | 127.3663 |
| Ear11 | Eosinophil-associated, ribonuclease A family, member 11 | 96.4100 |
| Ear10 | Eosinophil-associated, ribonuclease A family, member 10 | 88.2232 |
| Cst7 | Cystatin F (leukocystatin) | 86.1656 |
| Tuba8 | Tubulin, α 8 | 58.8395 |
| Ear12 | Eosinophil-associated, ribonuclease A family, member 12 | 46.9911 |
| Asns | Asparagine synthetase | 46.9515 |
| Chi3l3 | Chitinase-like 3 or Ym1 | 44.8807 |
| Cdh1 | Cadherin 1 | 44.7643 |
| Cish | Cytokine inducible SH2-containing protein | 33.5730 |
| Gm4610 | Predicted gene 4610 | 28.9111 |
| Gm6756 | Predicted gene 6756 | 28.5975 |
| Gfra2 | Glial cell line derived neurotrophic factor family receptor α 2 | 28.3951 |
| Phgdh | 3-Phosphoglycerate dehydrogenase | 28.1069 |
| Slc7a2 | Solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 | 27.6141 |
| Gm5150 | Predicted gene 5150 | 26.6933 |
| A530064D06Rik | RIKEN cDNA A530064D06 gene | 26.5447 |
| Ear2 | Eosinophil-associated, ribonuclease A family, member 2 | 25.9995 |
| Gatm | Glycine amidinotransferase (l-arginine:glycine amidinotransferase) | 25.6794 |
| Pi16 | Peptidase inhibitor 16 | 23.3797 |
| Flt1 | FMS-like tyrosine kinase 1 | 23.0790 |
| Fbp1 | Fructose bisphosphatase 1 | 22.4783 |
| Apoe | Apolipoprotein E | 0.0499 |
| Clec4d | C-type lectin domain family 4, member d | 0.0532 |
| Apoc2 | Apolipoprotein C-II | 0.0551 |
| Atf7 | Activating transcription factor 7 | 0.0663 |
| Irf8 | Interferon regulatory factor 8 | 0.0682 |
| Gm11538 | Predicted gene 11538 | 0.0778 |
| Fcgr1 | Fc receptor, IgG, high affinity I | 0.0858 |
| Rgs1 | Regulator of G-protein signaling 1 | 0.0877 |
| Ccl3 | Chemokine (C-C motif) ligand 3 | 0.0880 |
| Prom1 | Prominin 1 | 0.0982 |
| Gm1673 | Predicted gene 1673 | 0.1016 |
| Cdkn1c | Cyclin-dependent kinase inhibitor 1C (P57) | 0.1045 |
| Olfr871 | Olfactory receptor 871 | 0.1055 |
| Neurl3 | Neuralized homolog 3 homolog (Drosophila) | 0.1063 |
| Lyz2 | Lysozyme 2 | 0.1138 |
| Itgb5 | Integrin β 5 | 0.1160 |
| Lst1 | Leukocyte specific transcript 1 | 0.1253 |
| Rnf150 | Ring finger protein 150 | 0.1274 |
| Itga6 | Integrin α 6 | 0.1277 |
| Cnrip1 | Cannabinoid receptor interacting protein 1 | 0.1286 |
| Abca1 | ATP-binding cassette, sub-family A (ABC1), member 1 | 0.1294 |
| Cbr2 | Carbonyl reductase 2 | 0.1316 |
| 1700003F12Rik | RIKEN cDNA 1700003F12 gene | 0.1339 |
| Lmo2 | LIM domain only 2 | 0.1416 |
| Ldhb | Lactate dehydrogenase B | 0.1431 |
| Gene symbol . | Description . | Fold change* . |
|---|---|---|
| AA467197 | Expressed sequence AA467197 | 367.4197 |
| Chi3l4 | Chitinase-like 4 or Ym2 | 310.8889 |
| Aqp3 | Aquaporin 3 | 156.1061 |
| Ddx4 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 | 127.3663 |
| Ear11 | Eosinophil-associated, ribonuclease A family, member 11 | 96.4100 |
| Ear10 | Eosinophil-associated, ribonuclease A family, member 10 | 88.2232 |
| Cst7 | Cystatin F (leukocystatin) | 86.1656 |
| Tuba8 | Tubulin, α 8 | 58.8395 |
| Ear12 | Eosinophil-associated, ribonuclease A family, member 12 | 46.9911 |
| Asns | Asparagine synthetase | 46.9515 |
| Chi3l3 | Chitinase-like 3 or Ym1 | 44.8807 |
| Cdh1 | Cadherin 1 | 44.7643 |
| Cish | Cytokine inducible SH2-containing protein | 33.5730 |
| Gm4610 | Predicted gene 4610 | 28.9111 |
| Gm6756 | Predicted gene 6756 | 28.5975 |
| Gfra2 | Glial cell line derived neurotrophic factor family receptor α 2 | 28.3951 |
| Phgdh | 3-Phosphoglycerate dehydrogenase | 28.1069 |
| Slc7a2 | Solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 | 27.6141 |
| Gm5150 | Predicted gene 5150 | 26.6933 |
| A530064D06Rik | RIKEN cDNA A530064D06 gene | 26.5447 |
| Ear2 | Eosinophil-associated, ribonuclease A family, member 2 | 25.9995 |
| Gatm | Glycine amidinotransferase (l-arginine:glycine amidinotransferase) | 25.6794 |
| Pi16 | Peptidase inhibitor 16 | 23.3797 |
| Flt1 | FMS-like tyrosine kinase 1 | 23.0790 |
| Fbp1 | Fructose bisphosphatase 1 | 22.4783 |
| Apoe | Apolipoprotein E | 0.0499 |
| Clec4d | C-type lectin domain family 4, member d | 0.0532 |
| Apoc2 | Apolipoprotein C-II | 0.0551 |
| Atf7 | Activating transcription factor 7 | 0.0663 |
| Irf8 | Interferon regulatory factor 8 | 0.0682 |
| Gm11538 | Predicted gene 11538 | 0.0778 |
| Fcgr1 | Fc receptor, IgG, high affinity I | 0.0858 |
| Rgs1 | Regulator of G-protein signaling 1 | 0.0877 |
| Ccl3 | Chemokine (C-C motif) ligand 3 | 0.0880 |
| Prom1 | Prominin 1 | 0.0982 |
| Gm1673 | Predicted gene 1673 | 0.1016 |
| Cdkn1c | Cyclin-dependent kinase inhibitor 1C (P57) | 0.1045 |
| Olfr871 | Olfactory receptor 871 | 0.1055 |
| Neurl3 | Neuralized homolog 3 homolog (Drosophila) | 0.1063 |
| Lyz2 | Lysozyme 2 | 0.1138 |
| Itgb5 | Integrin β 5 | 0.1160 |
| Lst1 | Leukocyte specific transcript 1 | 0.1253 |
| Rnf150 | Ring finger protein 150 | 0.1274 |
| Itga6 | Integrin α 6 | 0.1277 |
| Cnrip1 | Cannabinoid receptor interacting protein 1 | 0.1286 |
| Abca1 | ATP-binding cassette, sub-family A (ABC1), member 1 | 0.1294 |
| Cbr2 | Carbonyl reductase 2 | 0.1316 |
| 1700003F12Rik | RIKEN cDNA 1700003F12 gene | 0.1339 |
| Lmo2 | LIM domain only 2 | 0.1416 |
| Ldhb | Lactate dehydrogenase B | 0.1431 |
Calculated as average fold change of genes regulated by IL-4 in resident macrophage and genes regulated by IL-4 in monocyte-derived macrophages.
GO analysis of transcriptional profiles from monocyte-derived and tissue resident macrophage-derived AAMs. (A) Biological processes (BPs) induced in IL-4c expanded AAM (IL-4c) relative to resident macrophages (Res). x-axis indicates the amount of statistical significance [as −log(P)] in enrichment for the indicated biological process. Arrows depict pathways of interests mentioned in the text. (B) BPs induced in Thio+IL-4c-induced AAMs relative to Thio-induced macrophages. (C) BPs induced in both monocyte- and tissue-derived AAMs. (D) Molecular function (MF) pathways induced in Thio+IL-4c-induced AAMs relative to Thio-induced macrophages. (E) Hierarchical clustering analysis of 26 unsupervised k-means clusters of genes that have similar expression profiles. The averaged expression for all the genes in each cluster is shown as a single row. The biological significance of each cluster was determined by measuring the GO term enrichment for BPs. Results shown are for individual mice.
GO analysis of transcriptional profiles from monocyte-derived and tissue resident macrophage-derived AAMs. (A) Biological processes (BPs) induced in IL-4c expanded AAM (IL-4c) relative to resident macrophages (Res). x-axis indicates the amount of statistical significance [as −log(P)] in enrichment for the indicated biological process. Arrows depict pathways of interests mentioned in the text. (B) BPs induced in Thio+IL-4c-induced AAMs relative to Thio-induced macrophages. (C) BPs induced in both monocyte- and tissue-derived AAMs. (D) Molecular function (MF) pathways induced in Thio+IL-4c-induced AAMs relative to Thio-induced macrophages. (E) Hierarchical clustering analysis of 26 unsupervised k-means clusters of genes that have similar expression profiles. The averaged expression for all the genes in each cluster is shown as a single row. The biological significance of each cluster was determined by measuring the GO term enrichment for BPs. Results shown are for individual mice.
Genes up-regulated with IL-4 in monocyte-derived macrophages and identified by GO analysis of molecular function to be involved with chemokine activities
| Gene symbol . | Name . | Fold change . |
|---|---|---|
| Ccl17 | Chemokine (C-C motif) ligand 17 | 19.2074 |
| Gm5150 | Predicted gene 5150 | 4.1190 |
| Ccl2 | Chemokine (C-C motif) ligand 2 | 7.4465 |
| Ccl7 | Chemokine (C-C motif) ligand 7 | 21.4119 |
| Ccl12 | Chemokine (C-C motif) ligand 12 | 7.4850 |
| Ccl24 | Chemokine (C-C motif) ligand 24 | 308.1040 |
| Gene symbol . | Name . | Fold change . |
|---|---|---|
| Ccl17 | Chemokine (C-C motif) ligand 17 | 19.2074 |
| Gm5150 | Predicted gene 5150 | 4.1190 |
| Ccl2 | Chemokine (C-C motif) ligand 2 | 7.4465 |
| Ccl7 | Chemokine (C-C motif) ligand 7 | 21.4119 |
| Ccl12 | Chemokine (C-C motif) ligand 12 | 7.4850 |
| Ccl24 | Chemokine (C-C motif) ligand 24 | 308.1040 |
Genes down-regulated with IL-4 in resident macrophages and identified by GO analysis of molecular function to be involved with chemokine activities
| Gene symbol . | Name . | Fold change . |
|---|---|---|
| Cxcl13 | Chemokine (C-X-C motif) ligand 13 | 0.0020 |
| Ccl3 | Chemokine (C-C motif) ligand 3 | 0.0439 |
| Cxcl2 | Chemokine (C-X-C motif) ligand 2 | 0.1496 |
| Cxcl1 | Chemokine (C-X-C motif) ligand 1 | 0.1512 |
| Gene symbol . | Name . | Fold change . |
|---|---|---|
| Cxcl13 | Chemokine (C-X-C motif) ligand 13 | 0.0020 |
| Ccl3 | Chemokine (C-C motif) ligand 3 | 0.0439 |
| Cxcl2 | Chemokine (C-X-C motif) ligand 2 | 0.1496 |
| Cxcl1 | Chemokine (C-X-C motif) ligand 1 | 0.1512 |
Genes up-regulated with IL-4 in monocyte-derived macrophages and identified by GO analysis of molecular function to be involved with cytokine activities
| Gene symbol . | Name . | Fold change . |
|---|---|---|
| Gm5150 | Predicted gene 5150 | 4.1190 |
| Ccl12 | Chemokine (C-C motif) ligand 12 | 7.4850 |
| Ccl7 | Chemokine (C-C motif) ligand 7 | 21.4119 |
| Il6 | Interleukin 6 | 8.8808 |
| Ccl2 | Chemokine (C-C motif) ligand 2 | 6.9844 |
| Cish | Cytokine inducible SH2-containing protein | 59.3660 |
| C3 | Complement component 3 | 9.2433 |
| Il31ra | Interleukin 31 receptor A | 9.6304 |
| Ccl24 | Chemokine (C-C motif) ligand 24 | 308.1040 |
| Socs6 | Suppressor of cytokine signaling 6 | 3.3646 |
| Ccl17 | Chemokine (C-C motif) ligand 17 | 19.2074 |
| Socs2 | Suppressor of cytokine signaling 2 | 7.4007 |
| Gene symbol . | Name . | Fold change . |
|---|---|---|
| Gm5150 | Predicted gene 5150 | 4.1190 |
| Ccl12 | Chemokine (C-C motif) ligand 12 | 7.4850 |
| Ccl7 | Chemokine (C-C motif) ligand 7 | 21.4119 |
| Il6 | Interleukin 6 | 8.8808 |
| Ccl2 | Chemokine (C-C motif) ligand 2 | 6.9844 |
| Cish | Cytokine inducible SH2-containing protein | 59.3660 |
| C3 | Complement component 3 | 9.2433 |
| Il31ra | Interleukin 31 receptor A | 9.6304 |
| Ccl24 | Chemokine (C-C motif) ligand 24 | 308.1040 |
| Socs6 | Suppressor of cytokine signaling 6 | 3.3646 |
| Ccl17 | Chemokine (C-C motif) ligand 17 | 19.2074 |
| Socs2 | Suppressor of cytokine signaling 2 | 7.4007 |
List of top 50 genes down-regulated by IL-4 in resident macrophages
| Gene symbol . | Name . | Fold change . |
|---|---|---|
| Cxcl13 | Chemokine (C-X-C motif) ligand 13 | 0.0020 |
| Lyz2 | Lysozyme 2 | 0.0081 |
| Atf7 | Activating transcription factor 7 | 0.0131 |
| Lpl | Lipoprotein lipase | 0.0177 |
| Irf8 | Interferon regulatory factor 8 | 0.0192 |
| Apoc2 | Apolipoprotein C-II | 0.0196 |
| Dusp6 | Dual specificity phosphatase 6 | 0.0219 |
| Clec7a | C-type lectin domain family 7, member a | 0.0238 |
| Apoc1 | Apolipoprotein C-I | 0.0300 |
| Hp | Haptoglobin | 0.0339 |
| Gm1673 | Predicted gene 1673 | 0.0344 |
| Gm11538 | Predicted gene 11538 | 0.0367 |
| Fcgr1 | Fc receptor, IgG, high affinity I | 0.0375 |
| Pltp | Phospholipid transfer protein | 0.0386 |
| Ccl3 | Chemokine (C-C motif) ligand 3 | 0.0439 |
| Abca1 | ATP-binding cassette, sub-family A (ABC1), member 1 | 0.0441 |
| Hspa1a | Heat shock protein 1A | 0.0494 |
| Rgs2 | Regulator of G-protein signaling 2 | 0.0526 |
| Lama3 | Laminin, α 3 | 0.0563 |
| Nat8l | N-acetyltransferase 8-like | 0.0564 |
| Apoc4 | Apolipoprotein C-IV | 0.0644 |
| Clec4d | C-type lectin domain family 4, member d | 0.0673 |
| Rab6b | RAB6B, member RAS oncogene family | 0.0704 |
| Emb | Embigin | 0.0707 |
| F13a1 | Coagulation factor XIII, A1 subunit | 0.0713 |
| Fpr1 | Formyl peptide receptor 1 | 0.0714 |
| Upk1a | Uroplakin 1A | 0.0718 |
| Cd63 | CD63 antigen | 0.0744 |
| Gm4788 | Predicted gene 4788 | 0.0773 |
| Ccm2l | Cerebral cavernous malformation 2-like | 0.0782 |
| Marco | Macrophage receptor with collagenous structure | 0.0804 |
| Fpr2 | Formyl peptide receptor 2 | 0.0805 |
| Fkbp1a | FK506 binding protein 1a | 0.0805 |
| 1700003F12Rik | RIKEN cDNA 1700003F12 gene | 0.0806 |
| Apoe | Apolipoprotein E | 0.0850 |
| Rprm | Reprimo, TP53 dependent G2 arrest mediator candidate | 0.0854 |
| Cfhr2 | Complement factor H-related 2 | 0.0855 |
| Abca9 | ATP-binding cassette, sub-family A (ABC1), member 9 | 0.0913 |
| Cfh | Complement component factor h | 0.0924 |
| Egfl7 | EGF-like domain 7 | 0.0930 |
| Olfr871 | Olfactory receptor 871 | 0.0934 |
| Neurl3 | Neuralized homolog 3 homolog (Drosophila) | 0.0949 |
| Rgs1 | Regulator of G-protein signaling 1 | 0.0949 |
| Rs5-8s1 | Unknown | 0.0977 |
| Prom1 | Prominin 1 | 0.0986 |
| Ldhb | Lactate dehydrogenase B | 0.0987 |
| Rhbdf1 | Rhomboid family 1 (Drosophila) | 0.1027 |
| Fut7 | Fucosyltransferase 7 | 0.1038 |
| Pilra | Paired immunoglobin-like type 2 receptor α | 0.1082 |
| Lst1 | Leukocyte specific transcript 1 | 0.1086 |
| Gene symbol . | Name . | Fold change . |
|---|---|---|
| Cxcl13 | Chemokine (C-X-C motif) ligand 13 | 0.0020 |
| Lyz2 | Lysozyme 2 | 0.0081 |
| Atf7 | Activating transcription factor 7 | 0.0131 |
| Lpl | Lipoprotein lipase | 0.0177 |
| Irf8 | Interferon regulatory factor 8 | 0.0192 |
| Apoc2 | Apolipoprotein C-II | 0.0196 |
| Dusp6 | Dual specificity phosphatase 6 | 0.0219 |
| Clec7a | C-type lectin domain family 7, member a | 0.0238 |
| Apoc1 | Apolipoprotein C-I | 0.0300 |
| Hp | Haptoglobin | 0.0339 |
| Gm1673 | Predicted gene 1673 | 0.0344 |
| Gm11538 | Predicted gene 11538 | 0.0367 |
| Fcgr1 | Fc receptor, IgG, high affinity I | 0.0375 |
| Pltp | Phospholipid transfer protein | 0.0386 |
| Ccl3 | Chemokine (C-C motif) ligand 3 | 0.0439 |
| Abca1 | ATP-binding cassette, sub-family A (ABC1), member 1 | 0.0441 |
| Hspa1a | Heat shock protein 1A | 0.0494 |
| Rgs2 | Regulator of G-protein signaling 2 | 0.0526 |
| Lama3 | Laminin, α 3 | 0.0563 |
| Nat8l | N-acetyltransferase 8-like | 0.0564 |
| Apoc4 | Apolipoprotein C-IV | 0.0644 |
| Clec4d | C-type lectin domain family 4, member d | 0.0673 |
| Rab6b | RAB6B, member RAS oncogene family | 0.0704 |
| Emb | Embigin | 0.0707 |
| F13a1 | Coagulation factor XIII, A1 subunit | 0.0713 |
| Fpr1 | Formyl peptide receptor 1 | 0.0714 |
| Upk1a | Uroplakin 1A | 0.0718 |
| Cd63 | CD63 antigen | 0.0744 |
| Gm4788 | Predicted gene 4788 | 0.0773 |
| Ccm2l | Cerebral cavernous malformation 2-like | 0.0782 |
| Marco | Macrophage receptor with collagenous structure | 0.0804 |
| Fpr2 | Formyl peptide receptor 2 | 0.0805 |
| Fkbp1a | FK506 binding protein 1a | 0.0805 |
| 1700003F12Rik | RIKEN cDNA 1700003F12 gene | 0.0806 |
| Apoe | Apolipoprotein E | 0.0850 |
| Rprm | Reprimo, TP53 dependent G2 arrest mediator candidate | 0.0854 |
| Cfhr2 | Complement factor H-related 2 | 0.0855 |
| Abca9 | ATP-binding cassette, sub-family A (ABC1), member 9 | 0.0913 |
| Cfh | Complement component factor h | 0.0924 |
| Egfl7 | EGF-like domain 7 | 0.0930 |
| Olfr871 | Olfactory receptor 871 | 0.0934 |
| Neurl3 | Neuralized homolog 3 homolog (Drosophila) | 0.0949 |
| Rgs1 | Regulator of G-protein signaling 1 | 0.0949 |
| Rs5-8s1 | Unknown | 0.0977 |
| Prom1 | Prominin 1 | 0.0986 |
| Ldhb | Lactate dehydrogenase B | 0.0987 |
| Rhbdf1 | Rhomboid family 1 (Drosophila) | 0.1027 |
| Fut7 | Fucosyltransferase 7 | 0.1038 |
| Pilra | Paired immunoglobin-like type 2 receptor α | 0.1082 |
| Lst1 | Leukocyte specific transcript 1 | 0.1086 |
List of top 50 genes downregulated by IL-4 in monocyte-derived macrophages
| Gene symbol . | Name . | Fold change . |
|---|---|---|
| Mamdc2 | MAM domain containing 2 | 0.0148 |
| Apoe | Apolipoprotein E | 0.0148 |
| Dio2 | Deiodinase, iodothyronine, type II | 0.0169 |
| AF251705 | cDNA sequence AF251705 | 0.0211 |
| Cdkn1c | Cyclin-dependent kinase inhibitor 1C (P57) | 0.0226 |
| Hpgds | Hematopoietic prostaglandin D synthase | 0.0232 |
| Syt10 | Synaptotagmin X | 0.0287 |
| C77080 | Expressed sequence C77080 | 0.0338 |
| Mmp27 | Matrix metallopeptidase 27 | 0.0356 |
| Mcoln3 | Mucolipin 3 | 0.0364 |
| Tspan7 | Tetraspanin 7 | 0.0368 |
| Htra1 | HtrA serine peptidase 1 | 0.0371 |
| Saa3 | Serum amyloid A 3 | 0.0373 |
| Clec4d | C-type lectin domain family 4, member d | 0.0390 |
| Cd200 | CD200 antigen | 0.0412 |
| Ms4a7 | Membrane-spanning 4-domains, subfamily A, member 7 | 0.0413 |
| Sh2d3c | SH2 domain containing 3C | 0.0430 |
| Htr2b | 5-Hydroxytryptamine (serotonin) receptor 2B | 0.0452 |
| Reps2 | RALBP1 associated Eps domain containing protein 2 | 0.0489 |
| Npy | Neuropeptide Y | 0.0494 |
| Cnrip1 | Cannabinoid receptor interacting protein 1 | 0.0495 |
| Tmem140 | Transmembrane protein 140 | 0.0502 |
| Lhfpl2 | Lipoma HMGIC fusion partner-like 2 | 0.0503 |
| Arhgap22 | ρ GTPase activating protein 22 | 0.0507 |
| Prss46 | Protease, serine 46 | 0.0512 |
| Spp1 | Secreted phosphoprotein 1 | 0.0513 |
| Cd93 | CD93 antigen | 0.0519 |
| Pcp4l1 | Purkinje cell protein 4-like 1 | 0.0525 |
| Mfge8 | Milk fat globule-EGF factor 8 protein | 0.0547 |
| Gstm1 | Glutathione S-transferase, μ 1 | 0.0547 |
| Gpr137b | G protein-coupled receptor 137B | 0.0576 |
| Cd109 | CD109 antigen | 0.0581 |
| Tspan13 | Tetraspanin 13 | 0.0586 |
| Gas6 | Growth arrest specific 6 | 0.0587 |
| 4930404N11Rik | RIKEN cDNA 4930404N11 gene | 0.0631 |
| Mpeg1 | Macrophage expressed gene 1 | 0.0655 |
| Tecpr1 | Tectonin β-propeller repeat containing 1 | 0.0680 |
| Gprc5b | G protein-coupled receptor, family C, group 5, member B | 0.0703 |
| Pianp | PILR α associated neural protein | 0.0707 |
| Gpr137b-ps | G protein-coupled receptor 137B, pseudogene | 0.0734 |
| Gdpd1 | Glycerophosphodiester phosphodiesterase domain containing 1 | 0.0742 |
| Lrrc27 | Leucine rich repeat containing 27 | 0.0748 |
| Fstl1 | Follistatin-like 1 | 0.0750 |
| Pdzk1ip1 | PDZK1 interacting protein 1 | 0.0759 |
| Fhdc1 | FH2 domain containing 1 | 0.0765 |
| Itgb5 | Integrin β 5 | 0.0792 |
| Speg | SPEG complex locus | 0.0795 |
| Gstm3 | Glutathione S-transferase, μ 3 | 0.0799 |
| Rgs1 | Regulator of G-protein signaling 1 | 0.0805 |
| Gene symbol . | Name . | Fold change . |
|---|---|---|
| Mamdc2 | MAM domain containing 2 | 0.0148 |
| Apoe | Apolipoprotein E | 0.0148 |
| Dio2 | Deiodinase, iodothyronine, type II | 0.0169 |
| AF251705 | cDNA sequence AF251705 | 0.0211 |
| Cdkn1c | Cyclin-dependent kinase inhibitor 1C (P57) | 0.0226 |
| Hpgds | Hematopoietic prostaglandin D synthase | 0.0232 |
| Syt10 | Synaptotagmin X | 0.0287 |
| C77080 | Expressed sequence C77080 | 0.0338 |
| Mmp27 | Matrix metallopeptidase 27 | 0.0356 |
| Mcoln3 | Mucolipin 3 | 0.0364 |
| Tspan7 | Tetraspanin 7 | 0.0368 |
| Htra1 | HtrA serine peptidase 1 | 0.0371 |
| Saa3 | Serum amyloid A 3 | 0.0373 |
| Clec4d | C-type lectin domain family 4, member d | 0.0390 |
| Cd200 | CD200 antigen | 0.0412 |
| Ms4a7 | Membrane-spanning 4-domains, subfamily A, member 7 | 0.0413 |
| Sh2d3c | SH2 domain containing 3C | 0.0430 |
| Htr2b | 5-Hydroxytryptamine (serotonin) receptor 2B | 0.0452 |
| Reps2 | RALBP1 associated Eps domain containing protein 2 | 0.0489 |
| Npy | Neuropeptide Y | 0.0494 |
| Cnrip1 | Cannabinoid receptor interacting protein 1 | 0.0495 |
| Tmem140 | Transmembrane protein 140 | 0.0502 |
| Lhfpl2 | Lipoma HMGIC fusion partner-like 2 | 0.0503 |
| Arhgap22 | ρ GTPase activating protein 22 | 0.0507 |
| Prss46 | Protease, serine 46 | 0.0512 |
| Spp1 | Secreted phosphoprotein 1 | 0.0513 |
| Cd93 | CD93 antigen | 0.0519 |
| Pcp4l1 | Purkinje cell protein 4-like 1 | 0.0525 |
| Mfge8 | Milk fat globule-EGF factor 8 protein | 0.0547 |
| Gstm1 | Glutathione S-transferase, μ 1 | 0.0547 |
| Gpr137b | G protein-coupled receptor 137B | 0.0576 |
| Cd109 | CD109 antigen | 0.0581 |
| Tspan13 | Tetraspanin 13 | 0.0586 |
| Gas6 | Growth arrest specific 6 | 0.0587 |
| 4930404N11Rik | RIKEN cDNA 4930404N11 gene | 0.0631 |
| Mpeg1 | Macrophage expressed gene 1 | 0.0655 |
| Tecpr1 | Tectonin β-propeller repeat containing 1 | 0.0680 |
| Gprc5b | G protein-coupled receptor, family C, group 5, member B | 0.0703 |
| Pianp | PILR α associated neural protein | 0.0707 |
| Gpr137b-ps | G protein-coupled receptor 137B, pseudogene | 0.0734 |
| Gdpd1 | Glycerophosphodiester phosphodiesterase domain containing 1 | 0.0742 |
| Lrrc27 | Leucine rich repeat containing 27 | 0.0748 |
| Fstl1 | Follistatin-like 1 | 0.0750 |
| Pdzk1ip1 | PDZK1 interacting protein 1 | 0.0759 |
| Fhdc1 | FH2 domain containing 1 | 0.0765 |
| Itgb5 | Integrin β 5 | 0.0792 |
| Speg | SPEG complex locus | 0.0795 |
| Gstm3 | Glutathione S-transferase, μ 3 | 0.0799 |
| Rgs1 | Regulator of G-protein signaling 1 | 0.0805 |
To more systematically identify modularity in the expression data and potentially coregulated genes, we used unsupervised k-means clustering to identify clusters of genes that have similar expression profiles (Figure 3E; supplemental Figure 4). We identified 249 clusters based on optimal cluster fitting with the least complexity (supplemental Figure 4). Twenty-six clusters that had the most interesting expression patterns were selected for further investigation (Figure 3E). Figure 3E shows a hierarchical clustering analysis of the averaged expression for all the genes in each cluster shown as a single row. The biological significance of each cluster was determined by measuring the GO term enrichment for biological processes. Interestingly, genes involved in metabolic processes were highly enriched in AAMs induced by IL-4c alone. Genes involved in proliferation and cell cycle were enriched in both types of AAMs, reflecting the fact that they were both proliferating rapidly with a high proportion of cells in S phase (Figure 1A). These results indicate that monocyte-derived AAMs (Thio+IL-4c) and tissue macrophage-derived AAMs (IL-4c alone) have fundamentally different molecular signatures and that genes involved in metabolic processes are especially up-regulated in AAMs induced by IL-4c alone, and monocyte-derived AAMs up-regulate genes important in immune responses.
Key markers of alternative activation that are only expressed on monocyte-derived macrophages
The transcriptional analysis indicated that the message for PD-L2 (also known as B7-DC or Pdcd1lg2), a cell surface marker of alternative activation,22 was up-regulated by IL-4 on monocyte-derived macrophages but not resident peritoneal cells. This led us to evaluate cell surface expression of PD-L2, along with the mannose receptor CD206 (also known as MMR or Mrc1), the first and best known marker described for AAMs28 (gating strategy; supplemental Figure 5). F4/80int cells induced by Thio+IL-4c expressed both PD-L2 and CD206, whereas the majority of IL-4c-induced F4/80high tissue-derived AAMs did not (Figure 4A-B). CD206 was also expressed on Thio-elicited macrophages that are not alternatively activated, whereas PD-L2 was only expressed on Thio-elicited macrophages in response to IL-4 (Figure 4B). Up-regulation of PD-L2 was Stat-6 dependent, whereas CD206 was expressed independently of Stat-6 (Figure 4D-E). We also assessed MHC class II, which was up-regulated on monocyte-derived AAMs but not on F4/80high tissue-derived AAMs (Figure 4C). The minor F4/80int population in naïve and IL-4c alone-treated mice expressed CD206, and only this minor subset expressed PD-L2 and MHC class II after exposure to IL-4c (Figure 4A-C). These cells were also CX3CR1-GFP+ (Figure 1G) and are phenotypically consistent with a monocyte-derived origin.25 This indicates that the differences in phenotype we observe are not unique to thioglycollate injection but are a general feature of monocyte-derived macrophages.
AAMs derived from inflammatory monocytes express PD-L2, CD206, and MHC class II, but not tissue macrophage-derived AAMs. FACS analysis of (A) PD-L2, (B) CD206 and (C) MHC class II from peritoneal macrophages of mice untreated (naïve/resident) or injected i.p. with IL-4c alone (IL-4c) or thioglycollate alone (Thio) or Thio and IL-4c (Thio+IL-4c). Graphs depict the geometric median fluorescent intensity (MFI) of PD-L2 and CD206 and percentage of MHC class II, gated on CD11b+ cells from the peritoneal cavity of individual mice. Data are representative of 3 independent experiments. (D) Stat6 is required for regulating PD-L2 but not CD206 expression in monocyte-derived AAMs. (E) Quantitation of CD11b+ cells that are CD206+PD-L2+ from the peritoneal cavity of individual mice and (F) the total number of peritoneal cavity cells recovered from treated animals. Results shown are representative of 2 independent experiments. *P < .05 and **P < .01 as determined by ANOVA.
AAMs derived from inflammatory monocytes express PD-L2, CD206, and MHC class II, but not tissue macrophage-derived AAMs. FACS analysis of (A) PD-L2, (B) CD206 and (C) MHC class II from peritoneal macrophages of mice untreated (naïve/resident) or injected i.p. with IL-4c alone (IL-4c) or thioglycollate alone (Thio) or Thio and IL-4c (Thio+IL-4c). Graphs depict the geometric median fluorescent intensity (MFI) of PD-L2 and CD206 and percentage of MHC class II, gated on CD11b+ cells from the peritoneal cavity of individual mice. Data are representative of 3 independent experiments. (D) Stat6 is required for regulating PD-L2 but not CD206 expression in monocyte-derived AAMs. (E) Quantitation of CD11b+ cells that are CD206+PD-L2+ from the peritoneal cavity of individual mice and (F) the total number of peritoneal cavity cells recovered from treated animals. Results shown are representative of 2 independent experiments. *P < .05 and **P < .01 as determined by ANOVA.
To determine whether the differences we observed were physiologically relevant to infection, we chose 2 models that induce high levels of AAMs at the infection site but for which macrophage origins differed. Infection of mice with S mansoni leads to deposition of parasite eggs into the liver, with a large accumulation of AAMs in the granulomas surrounding the eggs. We previously demonstrated that AAMs induced by S mansoni infection are CX3CR1-GFPhigh,29 suggesting a monocyte origin for these cells. Recent monocyte transfer and intravital imaging studies have confirmed that these AAM are monocyte-derived (N.M.G., U.M.G., L.N.W., M. Carbrera, U. Frevert, and P.L., unpublished data, March 6, 2014). In contrast, infection with L sigmodontis leads to an expansion of AAMs in the pleural cavity that are almost entirely derived from the resident pool, as demonstrated by shielded bone marrow chimeras at 12 days after infection.11 We examined expression of PD-L2 and CD206 on AAMs induced by these 2 distinct helminth infections. AAMs induced by L sigmodontis adult worms residing in the pleural cavity did not express PD-L2 and CD206 (Figure 5A), the identical expression pattern to AAMs derived from IL-4c injection. In contrast, injection of Thio+IL-4c into the pleural cavity induced accumulation of PD-L2- and CD206- expressing AAMs (Figure 5A-B). As previously observed in the peritoneal cavity, macrophages in the pleural cavity induced by thioglycollate alone expressed CD206 but did not express PD-L2, which was only induced by Thio+IL-4c injection.
Different helminth infections induce either monocyte-derived or tissue-derived AAMs. (A) Representative FACs plots of pleural cavity macrophages on day 12 after L sigmodontis infection compared with pleural cavity macrophages of mice untreated (Res) or injected i.p. with thioglycollate alone (Thio) or Thio and IL-4c (Thio+IL-4c) or IL-4c alone (IL-4c). (B) Graphs depict proportion of CD206 and PD-L2 positive pleural cavity macrophages. (C) Representative FACs plots of PD-L2 and CD206 expression gated on CD11b+ cells isolated from the liver of S mansoni-infected mice (8 weeks after infection) compared with CD11b+ cells isolated from the liver of mice untreated (naïve) mice. (D) FIZZ1/RELMa intracellular staining on PD-L2+CD206+ macrophages from S mansoni-infected livers compared with PD-L2−CD206− macrophages. (E) Graph depicts the percentage of PD-L2 and CD206 double positive cells, gated on CD11b+ cells, from the livers of individual mice. (F) MFI of FIZZ1/RELMa staining on PD-L2+CD206+ and PD-L2−CD206− macrophages from livers of individual infected mice. *P < .05 and **P < .01 as determined by ANOVA. Results shown representative of 2 independent experiments.
Different helminth infections induce either monocyte-derived or tissue-derived AAMs. (A) Representative FACs plots of pleural cavity macrophages on day 12 after L sigmodontis infection compared with pleural cavity macrophages of mice untreated (Res) or injected i.p. with thioglycollate alone (Thio) or Thio and IL-4c (Thio+IL-4c) or IL-4c alone (IL-4c). (B) Graphs depict proportion of CD206 and PD-L2 positive pleural cavity macrophages. (C) Representative FACs plots of PD-L2 and CD206 expression gated on CD11b+ cells isolated from the liver of S mansoni-infected mice (8 weeks after infection) compared with CD11b+ cells isolated from the liver of mice untreated (naïve) mice. (D) FIZZ1/RELMa intracellular staining on PD-L2+CD206+ macrophages from S mansoni-infected livers compared with PD-L2−CD206− macrophages. (E) Graph depicts the percentage of PD-L2 and CD206 double positive cells, gated on CD11b+ cells, from the livers of individual mice. (F) MFI of FIZZ1/RELMa staining on PD-L2+CD206+ and PD-L2−CD206− macrophages from livers of individual infected mice. *P < .05 and **P < .01 as determined by ANOVA. Results shown representative of 2 independent experiments.
In contrast to L sigmodontis infection, we found that AAMs induced in the liver by S mansoni infection were similar in phenotype to AAMs induced by Thio+IL-4c into the peritoneal and pleural cavity. AAMs that accumulated in the liver granulomas induced by S mansoni eggs expressed PD-L2 and CD206 (Figure 5C,E). To confirm that PD-L2+ and CD206+ macrophages are AAMs, we also stained for intracellular FIZZ1/RELMa (Figure 5D,F) and gated on the PD-L2+CD206+ population compared with the PD-L2−CD206− population. The double positive cells clearly express more FIZZ1/RELMa than the negative compartment (Figure 5D,F). Thus, S mansoni and L sigmodontis infection, which induces AAMs from distinct origins, correspond with the phenotype of recruited or resident macrophage-derived cells observed following IL-4c injection into the peritoneal cavity.
AAMs derived from inflammatory monocytes can induce the differentiation of CD4+FoxP3+ cells through a retinoic acid-dependent mechanism
In a further effort to validate the microarray data, we also examined expression of the enzyme Raldh2 (or Aldh1a2). Raldh2 was chosen because we previously observed up-regulation in AAMs from the livers of S mansoni-infected mice,29 and it appeared to be specific to the monocyte-derived cells in our microarray analysis. Raldh2 is an enzyme that regulates production of retinoic acid (RA) with important implications for immune regulation.30 Raldh2 was up-regulated to a far greater extent by Thio+IL-4c than IL-4c only, suggesting that AAMs generated from monocytes preferentially produce RA (Figure 6A). We used the fluorescent substrate aldefluor (ALD) to confirm enzymatic activity by flow cytometry and found that Thio+IL-4c induced a substantial number of ALD+ cells that were F4/80int (Figure 6B; supplemental Figure 6). As expected from its monocyte origins, some of the minor F4/80int peritoneal population of naïve untreated animals and IL-4c-treated animals were also ALD+ (Figure 6C), although there were many fewer ALD+ cells than in Thio+IL-4c-treated animals.
Differential effects of monocyte- and tissue macrophage-derived AAMs on naïve CD4+ T cells. (A) RT-PCR analysis of Aldh1a2/Raldh2 expression in peritoneal macrophages normalized to expression of GAPDH. Graphs depict mean ± standard error of the mean of individual mice pooled from 5 to 6 independent experiments. (B) FACS analysis of Aldh activity gated on CD11b+ cells from the peritoneal cavity. Peritoneal cells were stained with aldefluor to detect Aldh activity for 2 hours prior to staining with cell surface antibodies antibodies. (C) Graph depicts the proportion of CD11b+ cells that are ALD+ from individual mice. Results are pooled from 4 independent experiments. (D) Flow cytometry contour plots showing the percentage of CD25+, Foxp3+CD4+ T cells after 6 days of coculture with peritoneal macrophages either with or without the RA inhibitor LE540 (1 µM). (E) Quantitation of the percentage of CD25+, Foxp3+ cells from the CD4+ compartment after coculture. Data are shown from 3 independent experiments. (F) Inhibition of CD4+ T-cell proliferation by IL-4c- and Thio+IL-4c-induced AAMs. FACS analysis of activated (anti-CD3+IL-2) Cell tracer-labeled naïve CD4+ cells cultured with peritoneal macrophages (ratio of 2:1) after 3 days of coculture. Results are representative of 3 independent experiments.
Differential effects of monocyte- and tissue macrophage-derived AAMs on naïve CD4+ T cells. (A) RT-PCR analysis of Aldh1a2/Raldh2 expression in peritoneal macrophages normalized to expression of GAPDH. Graphs depict mean ± standard error of the mean of individual mice pooled from 5 to 6 independent experiments. (B) FACS analysis of Aldh activity gated on CD11b+ cells from the peritoneal cavity. Peritoneal cells were stained with aldefluor to detect Aldh activity for 2 hours prior to staining with cell surface antibodies antibodies. (C) Graph depicts the proportion of CD11b+ cells that are ALD+ from individual mice. Results are pooled from 4 independent experiments. (D) Flow cytometry contour plots showing the percentage of CD25+, Foxp3+CD4+ T cells after 6 days of coculture with peritoneal macrophages either with or without the RA inhibitor LE540 (1 µM). (E) Quantitation of the percentage of CD25+, Foxp3+ cells from the CD4+ compartment after coculture. Data are shown from 3 independent experiments. (F) Inhibition of CD4+ T-cell proliferation by IL-4c- and Thio+IL-4c-induced AAMs. FACS analysis of activated (anti-CD3+IL-2) Cell tracer-labeled naïve CD4+ cells cultured with peritoneal macrophages (ratio of 2:1) after 3 days of coculture. Results are representative of 3 independent experiments.
Because RA can induce Foxp3 expression in CD4+ T cells, we compared the ability of IL-4c- and Thio+IL-4c-induced AAMs to induce expression of Foxp3 in naïve CD4+ T cells. Strikingly, only Thio+IL-4c induced AAMs were able to induce differentiation of Foxp3+ cells, detected after 7 days of culture (Figure 6D-E). Addition of the synthetic RA receptor antagonist LE540 blocked the induction of Foxp3+ expression by Thio+IL-4c-induced AAMs (Figure 6D), supporting a direct role for RA in the induction of Foxp3+ T-regulatory (Treg) cells by monocyte-derived AAMs. Importantly, tissue-derived AAMs did not induce Foxp3+ expression on CD4+ T cells, despite having the same ability to suppress T-cell proliferation (Figure 6F). Arginase activity has been shown to block T-cell proliferation,31,32 and because both types of AAMs express Arg1 at similarly high levels, this may contribute toward proliferative inhibition.
Uncoupling protein 1 is expressed only by resident macrophage-derived AAMs
Having confirmed specific features of monocyte-derived AAMs, we next validated potential markers for resident macrophage-derived AAMs. One of the most highly up-regulated genes was Ucp1 (Figure 7A), which is thought be highly selective for brown adipocytes,33 where it is responsible for thermogenesis.34 We confirmed by RT-PCR that Ucp1 was highly expressed by IL-4c treatment in peritoneal macrophages isolated by adherence (Figure 7B), as well as by FACS sorting on CD11b+F4/80+ cells (Figure 7C), but not expressed by Thio+IL-4c-induced AAMs. To confirm the expression of Ucp1 by resident-derived AAMs in a more physiological setting, we used the L sigmodontis infection model. AAMs induced in the pleural cavity of L sigmodontis-infected mice are resident cell derived11 and up-regulated Ucp1 expression (Figure 7D). Ucp1 expression increased in a linear fashion over 4 days after injection of IL-4c (Figure 7E) and was regulated in Stat6-dependent manner (Figure 7F). Finally, we confirmed that UCP1 protein was abundant in AAMs induced by IL-4c alone and was not detectable in Thio+IL-4c-induced AAMs (Figure 7G). Therefore, Ucp1 expression may prove a useful expression marker for tissue-derived AAMs.
Ucp1 is a unique marker for tissue resident macrophage-derived AAMs. (A) Microarray data for Ucp1 from AAMs. (B) Real-time PCR measurement of Ucp1 expression (relative to GAPDH) in peritoneal macrophages isolated by adherence. (C) RT-PCR measurement of Ucp1 expression in FACS sorted (CD11b+F4/80+) macrophages. (D) RT-PCR measurement of Ucp1 expression in pleural cavity macrophages on day 12 after L sigmodontis infection. (E) Time course analysis of Ucp1 expression after injection with IL-4c. (F) RT-PCR analysis of Ucp1 expression in peritoneal macrophages of treated Stat6-deficient animals. (G) Western blot analysis of Ucp1 protein from total peritoneal cells of individual mice treated with Thio, Thio+IL-4c, and IL-4c, with GAPDH as a loading control.
Ucp1 is a unique marker for tissue resident macrophage-derived AAMs. (A) Microarray data for Ucp1 from AAMs. (B) Real-time PCR measurement of Ucp1 expression (relative to GAPDH) in peritoneal macrophages isolated by adherence. (C) RT-PCR measurement of Ucp1 expression in FACS sorted (CD11b+F4/80+) macrophages. (D) RT-PCR measurement of Ucp1 expression in pleural cavity macrophages on day 12 after L sigmodontis infection. (E) Time course analysis of Ucp1 expression after injection with IL-4c. (F) RT-PCR analysis of Ucp1 expression in peritoneal macrophages of treated Stat6-deficient animals. (G) Western blot analysis of Ucp1 protein from total peritoneal cells of individual mice treated with Thio, Thio+IL-4c, and IL-4c, with GAPDH as a loading control.
Discussion
Tissue resident macrophages can be of embryonic origin and differ in cellular lineage from macrophages derived from monocytes that infiltrate tissues during an inflammatory response.5 In the context of helminth infection, IL-4/IL-13 can expand tissue resident AAMs11 or monocyte-derived AAMs,29,35 depending on the type of infection and the tissue inflicted. A key unanswered question is whether these 2 types of macrophages will respond differently to type 2 cytokines in ways that reflect their heterogeneous existing transcriptional programs and/or epigenetic differences.36 In this study, we find that AAMs from differing origins exhibit functional differences and cellular phenotypes that may reflect different physiological roles in type 2 responses. These findings have relevance beyond helminth infection because AAMs are now implicated in many noninfectious disease processes.37-39
This study also identifies potentially useful markers to indicate whether AAMs in a particular setting are derived from tissue-resident macrophages or from inflammatory monocytes. PD-L2 has been validated as a marker of AAM in several in vivo models of type 2 inflammation.23,35,40 Here, we show that IL-4 up-regulation of PD-L2 is limited to monocyte-derived macrophages. Further, PD-L2 expression is a more specific marker than the mannose receptor CD206,28 because thioglycollate-elicited macrophages also express CD206 in the absence of IL-4 and independently of Stat6. Hence, the absence of PD-L2 expression on macrophages that express Arg1, Relma, and Chi3l3 could be an indication of tissue-resident origin. In contrast, Ucp1 was thought to be highly selective to adipocytes,33 but we find here that it is highly expressed in tissue-derived AAMs. In the context of a type 2 immune response, Ucp1 may prove useful as a marker to distinguish between tissue-derived and monocyte-derived macrophages. The physiological relevance of Ucp1 expression by tissue-derived AAMs warrants further study.
The production of RA by dendritic cells (DCs) can drive gut-tropic immune responses and promote the differentiation of FoxP3+ Tregs.30 AAMs also express RALDH2, providing a source of RA that can induce the differentiation of FoxP3+ Tregs.29 The expression of RALDH2 was recently described in human AAMs.26 Here, we show that RALDH2 expression and functional ability to induce FoxP3+ Treg differentiation through RA is restricted to monocyte-derived AAMs. This restriction may reflect a greater need for Tregs during inflammatory responses involving monocyte recruitment. For example, the phagocytosis of apoptotic cells by inflammatory monocytes, instead of resident macrophages, results in a more inflammatory response,41 which may require Treg control. DCs rather than macrophages may be more anatomically positioned to induce Tregs in draining lymph nodes. Nonetheless, monocytes have recently been shown to traffic through the tissues into the lymph nodes.10 Perhaps monocyte-derived AAMs have more active roles in directing host responses than expanded resident AAMs. This would be consistent with up-regulated chemokine activity in monocyte-derived AAMs (Figure 3D) and down-regulation in tissue-derived AAMs (supplemental Figure 3). The preferential expression of MHC class II and thymus and activation regulated chemokine/Ccl17 by monocyte-derived AAMs (Table 4)42 is also notable because this may attract T cells to specifically interact with monocyte-derived AAMs.
The differences between global transcriptional profiles of tissue-derived AAMs vs monocyte-derived AAMs (Figure 2B) are greater than differences previously reported in the ImmGen project,43 perhaps because that study focused on tissue-resident macrophages. Molecules associated with antigen presentation, migration, and regulation of immune responses were preferentially associated with monocyte-derived AAMs, as was previously described for DCs.44 Because infiltrating monocytes during an inflammatory process can give rise to macrophages and DC-like cells depending on the environment, a key question is how they differ from the local resident macrophages and DCs as inflammation resolves. It is important to note that the interaction of thioglycollate with IL-4 is unclear and may synergistically enhance the observed immune signature on monocyte-derived AAMs. To exclude the effects of thioglycollate, future studies will examine the minor population of CX3CR1−GFP+PD-L2+ cells (Figure 4) induced by IL-4c treatment alone.
Egg granulomas from S mansoni infection predominantly recruit monocyte-derived AAMs, whereas L sigmodontis adult worms in the pleural cavity induce tissue-derived AAMs. A proximal explanation for this difference would be that the egg triggers the release of inflammatory chemokines (eg, Ccl2/MCP1) that recruit monocytes to a type 2 environment, whereas L sigmodontis infection does not. More fundamentally, are monocytes recruited to contain parasites, whereas resident cells minimize host damage? Although the answer is likely to be both tissue and parasite specific, it is not yet resolved whether inflammatory monocytes can eventually become tissue-resident F4/80-bright AAMs over time. In this study, we examine relatively short time periods after stimulation, but it will be important to know whether conversion occurs over time and whether the distinct functions we observed in this study are driven by cell origin or by long-term vs short-term tissue residence.
This article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Nikollaq Vozhilla for help with breeding and maintaining mice and Alison Fulton for maintenance of the L sigmodontis life cycle.
Flow cytometry was performed at the New York University Flow Cytometry and Cell Sorting Center, which is partially supported by National Institutes of Health (NIH), National Cancer Institute grant P30CA16087-31. This work is supported by NIH, National Institute of Allergy and Infectious Diseases grants AI093811 and AI094166 (P.L.), Ruth L. Kirschstein National Research Service Award fellowship F32AI102502 (N.M.G.), NIH National Cancer Institute training grant, T32 CA009161; Principle Investigator: Levy (U.B.R.), and Medical Research Council program grant MR/K01207X/1 (J.E.A.).
Authorship
Contribution: U.M.G. performed research and analyzed data; N.M.G. performed research and analyzed data; D.R. and S.J. performed research, analyzed data, and helped draft the manuscript; L.N.W. performed research; M.S.T. and Z.D.K. analyzed data; K.E.W. performed research; U.B.R. performed research; A.M. contributed vital reagents and designed research; and P.L. and J.E.A. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: P'ng Loke, Department of Microbiology, Division of Medical Parasitology, Old Public Health Building, Room 209, 341 East 25th St, New York, NY 10010; e-mail: png.loke@nyumc.org; or Judith E. Allen, Institute of Immunology and Infection Research, University of Edinburgh, Edinburgh EH9 3JT, United Kingdom; e-mail: j.allen@ed.ac.uk.
References
Author notes
U.M.G. and N.M.G. contributed equally to this work.

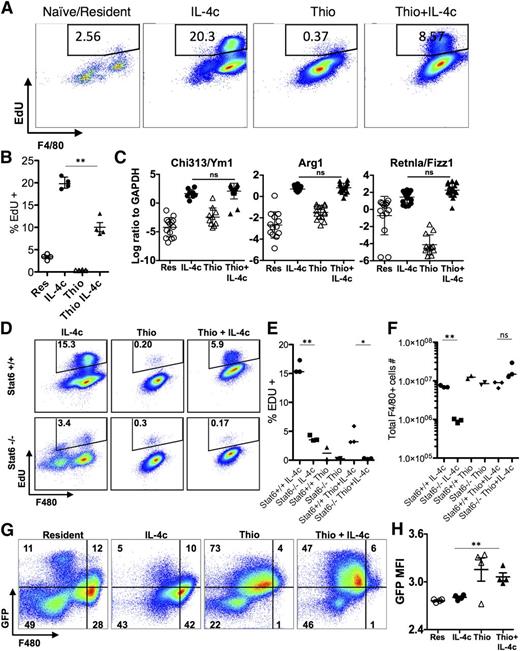
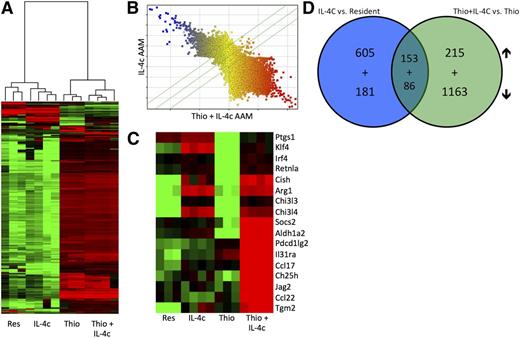
![Figure 3. GO analysis of transcriptional profiles from monocyte-derived and tissue resident macrophage-derived AAMs. (A) Biological processes (BPs) induced in IL-4c expanded AAM (IL-4c) relative to resident macrophages (Res). x-axis indicates the amount of statistical significance [as −log(P)] in enrichment for the indicated biological process. Arrows depict pathways of interests mentioned in the text. (B) BPs induced in Thio+IL-4c-induced AAMs relative to Thio-induced macrophages. (C) BPs induced in both monocyte- and tissue-derived AAMs. (D) Molecular function (MF) pathways induced in Thio+IL-4c-induced AAMs relative to Thio-induced macrophages. (E) Hierarchical clustering analysis of 26 unsupervised k-means clusters of genes that have similar expression profiles. The averaged expression for all the genes in each cluster is shown as a single row. The biological significance of each cluster was determined by measuring the GO term enrichment for BPs. Results shown are for individual mice.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/123/20/10.1182_blood-2013-08-520619/4/m_e110f3.jpeg?Expires=1771057177&Signature=pE6la6vQwcpKzzy8A7ueZK03jlQ9bSv89pBJILMNm802dAoLvaKIV-WtfnRJbX~50TrtLL8Z-arY2e3SBtoSW8uEipyQi~zvGPzGcCukMS67~1yRgPWFEQV4xQA7LfIxX15iE6MDlqF9oUkefzHJzkhVrjQ32~qZPUENd-ztcn5VTJ-M3fq01C550q-sCYe9WIFjkO9AVu7qyZSJeCLnhmRqQhKgz2bEcfkRrHpGMnCOkHdUWywDTuqBG6p50PdKNpY8UvPZW6t8QZeZt3fi9pjLrRjAq5ASu81KIDUhxO0kILNP55zauyDv4rB32ihadOgucVWd7yMPd0~e2nFQ1g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
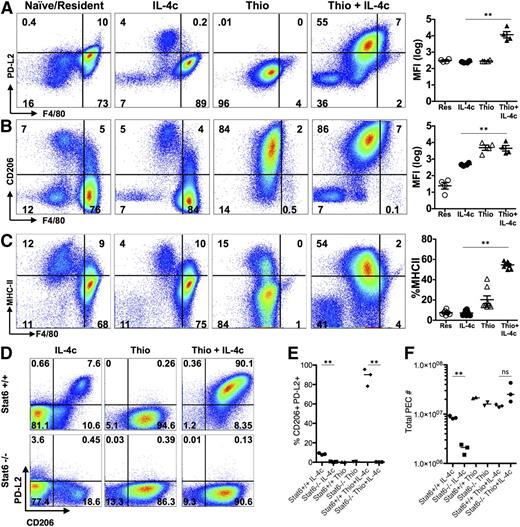
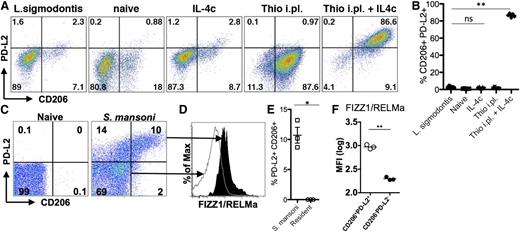
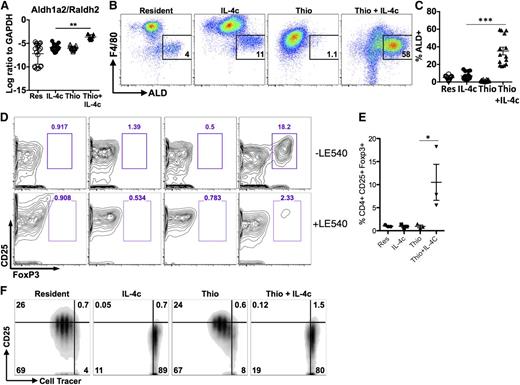
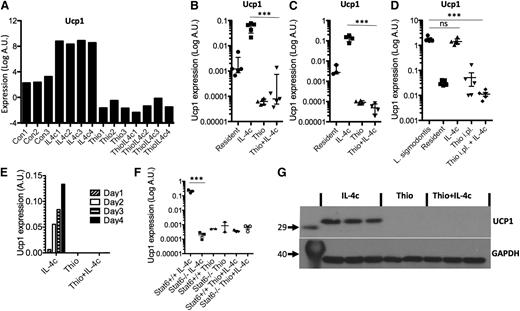
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal