Key Points
The high-resolution structure of the complex disulfide-bonded TIL′E′ (D′) region of VWF is presented.
The major factor VIII binding site is localized around a flexible region on the TIL′ domain.
Abstract
Although much of the function of von Willebrand factor (VWF) has been revealed, detailed insight into the molecular structure that enables VWF to orchestrate hemostatic processes, in particular factor VIII (FVIII) binding and stabilization in plasma, is lacking. Here, we present the high-resolution solution structure and structural dynamics of the D′ region of VWF, which constitutes the major FVIII binding site. D′ consists of 2 domains, trypsin-inhibitor–like (TIL′) and E′, of which the TIL′ domain lacks extensive secondary structure, is strikingly dynamic and harbors a cluster of pathological mutations leading to decreased FVIII binding affinity (type 2N von Willebrand disease [VWD]). This indicates that the backbone malleability of TIL′ is important for its biological activity. The principal FVIII binding site is localized to a flexible, positively charged region on TIL′, which is supported by the rigid scaffold of the TIL′ and E′ domain β sheets. Furthermore, surface-charge mapping of the TIL′E′ structure reveals a potential mechanism for the electrostatically guided, high-affinity VWF⋅FVIII interaction. Our findings provide novel insights into VWF⋅FVIII complex formation, leading to a greater understanding of the molecular basis of the bleeding diathesis type 2N VWD.
Introduction
The preservation of hemostatic integrity is secured by the activities of von Willebrand factor (VWF). Upon vascular damage, VWF acts as a molecular bridge facilitating the initial adhesion and aggregation of platelets to the site of vessel injury. Furthermore, VWF is the protective carrier of procoagulant factor VIII (FVIII) in plasma, thereby prolonging its half-life and efficiently localizing FVIII to the incipient platelet plug.1,2 The arrest of bleeding is critically dependent on VWF, as exemplified by von Willebrand disease (VWD), the most common inherited bleeding disorder in humans, which results from defective or deficient VWF protein.3
The importance of VWF⋅FVIII complex formation is illustrated by patients with severe VWD, who have undetectable VWF levels. Not only do these patients have a concomitant deficiency of FVIII, but they also have a considerably shortened survival of intravenously administered FVIII.1 This phenotype, which mimics hemophilia A, is also observed in patients with type 2N VWD, whose VWF harbors mutations that lead to decreased FVIII binding affinity.4 This behavior reflects the dependence of FVIII survival on the formation of VWF⋅FVIII complexes and illustrates the biological significance of this interaction. Hence, the importance of elucidating the structure of the FVIII binding region on VWF is 2-fold. First, it would provide insights into the mechanism of FVIII binding to VWF and reveal the minimal VWF unit required to stabilize FVIII. Second, analysis of VWD patient mutations against a background of both structural and functional perturbations will provide the link between genetic pathology and clinical phenotype.
Electron microscopy studies have given a first glimpse of the domain arrangement and overall structure of the multidomain VWF protein5 and in particular showed that the trypsin-inhibitor-like (TIL′) E′ domains form a protrusion from the D3 domain, thereby presenting the TIL′E′ domains to the physiological binding partner FVIII. However, the limited resolution of these structures has precluded the elucidation of the details needed to examine the molecular recognition and binding of FVIII to VWF. In addition, high-resolution structure determinations of the topologically complex disulfide-bonded VWF domains have not hitherto been feasible, except for the triplicated A domains,6,7 which are notable for their relative scarcity of cysteine residues. Thus, a detailed characterization of the interaction between VWF and FVIII has remained intractable, until now.
The FVIII binding region on VWF has been identified within a tryptic fragment termed SPIII-T4 (residues 767-1031).8 A recent domain assignment of VWF, based on conserved cysteine signatures,9 reveals that SPIII-T4 is composed of 3 distinct and conserved domains, namely TIL′ (residues 766-827), E′ (residues 829-863), and VWD3 (residues 867-1031). Accumulated lines of evidence suggest that of these regions it is the TIL′ and E′ domains (Figure 1), previously known collectively as D′, that are essential for FVIII binding.3 First, 72% of unique missense mutations that directly affect FVIII binding to VWF (type 2N VWD) are found in domains TIL′ and E′.10 Importantly, the most severe type 2N phenotypes, characterized by the lowest circulating plasma FVIII levels, are due to mutations in TIL′ (Figure 1 and supplemental Figure 1 available on the Blood Web site).4 Second, epitope-mapping studies of potent FVIII binding-blocking monoclonal antibodies (mAbs) mAb-418 (VWF epitope, residues 765-816) and mAb-32B12 (VWF epitope, residues 814-821) indicate that the putative FVIII binding site is localized to TIL′.11,12
Major FVIII binding sequence on VWF. The amino-terminal 99 residues (residues 766-864) of the mature VWF protein comprise the TIL′ and E′ domains, which form the basis of this paper. Positionally conserved cysteine residues are highlighted in black and numbered sequentially. Yellow lines show disulfide connectivities, and an asterisk marks connectivities that have been previously chemically assigned. The circled Cys846-863 (2-6) disulfide connectivity of the E′ domain is assigned in this study. Type 2N VWD mutations are shown with arrows directly under the sequence together with the 1-letter code of the mutation; type 2N mutations involving a cysteine residue are in black, and noncysteine mutations are in green. Secondary structure elements (arrow, β sheet; line, loop; spiral, 310 helix) are shown under the sequence.
Major FVIII binding sequence on VWF. The amino-terminal 99 residues (residues 766-864) of the mature VWF protein comprise the TIL′ and E′ domains, which form the basis of this paper. Positionally conserved cysteine residues are highlighted in black and numbered sequentially. Yellow lines show disulfide connectivities, and an asterisk marks connectivities that have been previously chemically assigned. The circled Cys846-863 (2-6) disulfide connectivity of the E′ domain is assigned in this study. Type 2N VWD mutations are shown with arrows directly under the sequence together with the 1-letter code of the mutation; type 2N mutations involving a cysteine residue are in black, and noncysteine mutations are in green. Secondary structure elements (arrow, β sheet; line, loop; spiral, 310 helix) are shown under the sequence.
Here, we present the solution structure and a characterization of the structural dynamics of the TIL′ and E′ domains of VWF. The TIL′E′ structure and dynamics provide an unprecedented view of the FVIII binding site on VWF and reveal a potential mechanism for the electrostatically guided VWF⋅FVIII interaction.
Materials and methods
Protein expression
The TIL′E′ coding sequence (residues 766-864) was amplified from full-length VWF complementary DNA and cloned into a pET32b+ vector containing an N-terminal thioredoxin (TRX) and His6 tag. The TRX-His6 tag was separated from the TIL′E′ coding sequence by a 31-residue linker (SSGLVPRGSGMKETAAAKFERQHMDSPDLGT) and a tobacco etch virus (TEV) cleavage sequence (ENLYFQ↓G), leaving residues GAMG as cloning artifacts. Constructs of TIL′E′ were expressed in Escherichia coli SHuffle cells13 and grown in ∼4 L of either lysogeny broth or M9 minimal medium. Isotope-labeled samples were grown in M9 minimal medium supplemented with 2.5 g/L 13C-glucose (uniform [U]-13C labeling) and 1 g/L 15NH4Cl (U-15N labeling) as the sole sources of carbon and nitrogen, respectively. For the 10% fractionally 13C-labeled TIL′E′ used for stereospecific assignment of the Val and Leu side-chain methyl groups, the M9 medium was supplemented with 10% U-13C-glucose and 90% unlabeled glucose. Cells were grown to an optical density (OD) at 600 nm of ∼0.6, and subsequent overexpression for 16 hours at 16°C was induced by the addition of 0.4 mM isopropyl-beta-D-thiogalactopyranoside.
Protein purification
After harvesting, the cells were resuspended in ∼40 mL of wash buffer (20 mM sodium phosphate, 500 mM NaCl, 40 mM imidazole [pH 7.4]). To the cell suspension was added 1 tablet of Complete Protease Inhibitor (Roche), nonionic and nondenaturing detergent in the form of 0.25% octylphenyl-polyethylene glycol, lysozyme, and DNase (5-10 μg/mL). Cells were broken by French press and subsequently centrifuged, and the cleared lysate (∼40 mL) was filtered (0.22 μm) and loaded onto a His-Trap Ni-nitrilotriacetic acid column (5 mL; GE Healthcare) pre-equilibrated with wash buffer. Bound protein was eluted using a linear gradient with increasing imidazole and pooled fractions dialyzed against 20 mM sodium phosphate, 100 mM NaCl (pH 7.4) for TEV proteolysis. TEV protease was added in the ratio of 1 OD280 unit TEV per 5 to 10 OD280 units of TRX-His6-TIL′E′; the reaction was incubated for 3 hours at room temperature and then at 4°C overnight to complete cleavage. Subsequently, the N-terminal TRX-His6 tag was separated from the cleaved TIL′E′ fraction on the His-Trap nitrilotriacetic acid column. The TIL′E′ was loaded onto a 16/60 Superdex 75 column (GE Healthcare) pre-equilibrated with 20 mM sodium phosphate, 100 mM NaCl (pH 7.4) as a final purification step. Monomeric TIL′E′ was concentrated to 450 μL with final sample concentrations of 0.2 to 0.4 mM for further analyses.
Isotope-labeled 15N and 13C,15N samples were prepared in 20 mM sodium phosphate, 100 mM NaCl, 10% D2O at pH 7.4 for nuclear magnetic resonance (NMR) spectroscopy. Samples used for backbone residual dipolar coupling (RDC) measurements were prepared by adding Pf1 bacteriophage (ASLA Biotech, Latvia)14 (∼5 mg/mL) or pentaethylene glycol dodecyl ether (C12E5)/n-hexanol (Sigma-Aldrich) (3% wt/wt C12E5, r = 0.94),15 where r is the molar ratio of C12E5 to n-hexanol, with corresponding deuterium quadrupolar splittings in Pf1 and C12E5/n-hexanol of ∼1.5 Hz and 8 Hz, respectively.
Mass spectrometry
Recombinant TIL′E′ (10 μM) was buffer exchanged into 100 mM ammonium acetate (pH 7.0) for mass spectrometry (MS) experiments, which were carried out on a Synapt HDMS (Waters) mass spectrometer.16 A 2.5-μL aliquot of the protein sample was delivered to the mass spectrometer by means of nanoelectrospray ionization using gold-coated capillaries (prepared in-house).
Ellman’s reagents
Formation of all disulfide bonds was verified by an Ellman’s assay with 5,5′-dithio-bis-(2-nitrobenzoic acid) which reacts with a free sulfhydryl group to yield 2-nitro-5-thiobenzoic acid, which was detected spectrophotometrically at 412 nm. The Ellman’s assay yielded ∼0.1 mol free -SH per mol TIL′E′, thus confirming that essentially all cysteine residues are engaged in disulfide-bond formation.
Binding of FVIII to plasma-derived VWF and recombinant TIL′E′ fragments
The potential FVIII binding partners (plasma-derived VWF, TIL′E′, reduced and alkylated TIL′E′) were coated directly onto a 96-well plate and incubated with an increasing concentration of FVIII. After equilibration and washing steps, bound FVIII was monitored with murine anti-FVIII and secondary anti-mouse horseradish peroxidase antibodies. Blanks were subtracted from the absorbance measurements, and 3 experiments were performed in duplicate. Details are given in supplemental Methods and materials.
NMR experiments
NMR experiments were recorded at 298 K on 500 MHz, 600 MHz, and 700 MHz spectrometers. Resonance assignments were obtained from a standard suite of triple-resonance 3-dimensional experiments (supplemental Methods and materials), and distance restraints were extracted from 15N- and 13C-edited nuclear Overhauser effect (NOE) spectroscopy spectra. RDCs were measured using aligned samples prepared in C12E5/n-hexanol15 (N-HN, HN-C′, N-C′, C-Cα, Cα-Cβ, and Cα-Hα) and Pf1 bacteriophage14 (N-HN). Backbone 15N relaxation rates (R1, R2, and {1H}15N steady-state heteronuclear NOEs) were measured at 600 MHz using standard methods17 and analyzed according to the Model-Free formalism.18 Relaxation dispersions were measured using a transverse relaxation optimized spectroscopy–selected Carr-Purcell-Meiboom-Gill (CPMG) experiment19 at 500 MHz and 700 MHz and analyzed in terms of a 2-site exchange model.
Structure calculations
The solution structure of TIL′E′ was calculated from the experimental NMR restraints using the programs CYANA20 and Xplor-NIH21 with the standard scripts provided. The structures shown are those from the Xplor-NIH calculation. In total, 954 unique distance restraints were obtained, of which 353 are long range (between residues that are >5 residues apart in the primary sequence); details are given in supplemental Methods and materials.
Results
Recombinantly produced TIL′E′ is correctly folded and disulfide bonded
Based on predictions of the FVIII binding site on VWF (Figure 1 and supplemental Figure 1), the TIL′ and E′ domains (residues 766-864), which are located just after the Furin cleavage site (residues 763↓764) in the primary sequence of VWF, were selected for structural characterization. Previous homology modeling9 predicted that all cysteine residues in TIL′E′ form disulfide bridges, of which 3 have been confirmed experimentally: Cys767-Cys808, Cys776-Cys804, and Cys810-Cys821 (Figure 1).22 Thus, isotopically labeled TIL′E′ for NMR structural and dynamic studies was obtained by overexpression in an E coli strain engineered to facilitate cytoplasmic disulfide-bond formation.13 Correct pairing of the 16 cysteine residues, and hence correct folding of TIL′E′, was critically dependent on the expression of these domains downstream of an N-terminal thioredoxin tag, yielding an initial translation product that bears resemblance to the native VWF prodomains, which contain oxidoreductase-like sequences.23
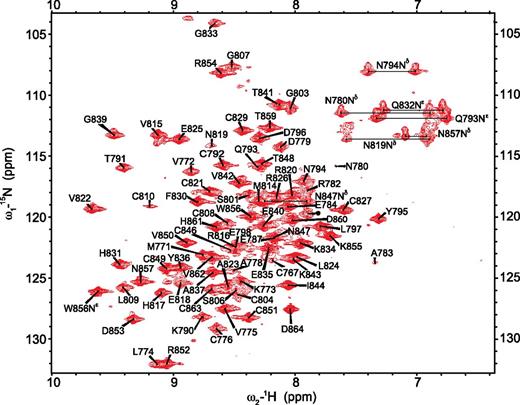
Recombinant TIL′E′ was analyzed by size-exclusion chromatography (supplemental Figure 2), electrospray-ionization MS (supplemental Figure 3), and Ellman’s assay, which together confirmed that TIL′E′ was homogeneous, monomeric, and contained a negligible amount of free thiol groups. Compelling evidence that the TIL′E′ protein is well folded is provided by the very narrow charge-state distribution of the native MS profile (supplemental Figure 3) and the wide 1HN chemical-shift dispersion in the 1H-15N heteronuclear single quantum coherence (HSQC) NMR spectrum (Figure 2). Closer inspection of the 1H-15N HSQC spectrum in Figure 2 revealed a heterogeneous distribution of peak intensities, providing the first indication that the TIL′ and E′ domains possess differential flexibility.
Assigned 1H-15N HSQC spectrum of VWF TIL′E′. The 1H-15N HSQC NMR spectrum of recombinant TIL′E′ measured at 25°C at 600 MHz. Each peak in this spectrum represents 1 backbone amide group, tryptophan side chain, or the Asn/Gln side chain. The chemical-shift assignments are annotated with the amino acid type and sequence number of the VWF protein. A near-complete backbone assignment of 1H-15N resonances is achieved. Of the 79 peaks observed, 77 backbone amides and the indole of Trp856 are assigned. A single unassigned peak in the center of the spectrum is indicated with an asterisk. Side-chain NH2 groups of Asn and Gln are assigned and shown connected by horizontal lines. There are broad peaks with lower intensity seen in the spectrum.
Assigned 1H-15N HSQC spectrum of VWF TIL′E′. The 1H-15N HSQC NMR spectrum of recombinant TIL′E′ measured at 25°C at 600 MHz. Each peak in this spectrum represents 1 backbone amide group, tryptophan side chain, or the Asn/Gln side chain. The chemical-shift assignments are annotated with the amino acid type and sequence number of the VWF protein. A near-complete backbone assignment of 1H-15N resonances is achieved. Of the 79 peaks observed, 77 backbone amides and the indole of Trp856 are assigned. A single unassigned peak in the center of the spectrum is indicated with an asterisk. Side-chain NH2 groups of Asn and Gln are assigned and shown connected by horizontal lines. There are broad peaks with lower intensity seen in the spectrum.
Recombinant TIL′E′ binds to FVIII and mAb-418
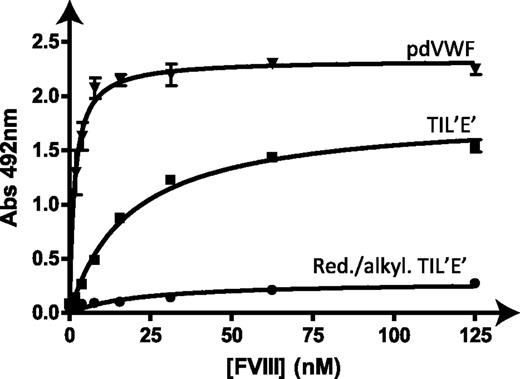
The best reporter of TIL′E′ function is binding to FVIII; this is dependent upon the folded TIL′E′ adopting its biologically active conformation. Solid-phase binding assays24 of the TIL′E′⋅FVIII interaction were undertaken to establish biological activity (Figure 3). Analysis of the binding assays showed that TIL′E′ binds to recombinant FVIII with an apparent affinity of 26 nM ± 2 nM, which is similar to the binding affinity of the SPIII-T4 fragment to FVIII (50-80 nM25,26 ). Reduced and alkylated TIL′E′ does not bind to FVIII, showing that the binding observed for TIL′E′ is due to specific rather than unspecific binding. Moreover, the binding affinity of the nonglycosylated TIL′E′ construct is at least as strong as the binding affinity of the slightly larger SPIII-T4 construct, demonstrating that glycosylation of TIL′E′ is not required for the specific TIL′E′⋅FVIII interaction. Using the same experimental setup, we also determined the binding affinity of plasma-derived VWF to FVIII, Kd = 0.9 nM, which is in the range of previously obtained results (eg, 0.52 nM,25 1.5 nM27 ), thus validating our result for recombinant TIL′E′. Binding of TIL′E′ to the conformation-dependent, FVIII binding-blocking VWF mAb-418 further confirmed the presence of a FVIII binding site on TIL′E′ (supplemental Figure 4). Overall, the binding experiments performed suggest that the recombinantly produced TIL′E′ is a faithful representation of these domains in full-length VWF and retains their biological function. Importantly, the binding of TIL′E′ to FVIII shown in Figure 3 identifies TIL′ and E′ as the domains of VWF that contribute the majority of the VWF⋅FVIII binding free energy.
Binding analysis of TIL′E′ functionality. Solid-phase binding assays of recombinant FVIII to plasma-derived VWF (pdVWF; ▾), TIL′E′ expressed in E coli. (TIL′E′; ▪), and reduced and alkylated TIL′E′ (Red./alkyl. TIL′E′; ●). The obtained apparent dissociation constants are pdVWF 0.9 nM ± 0.2 nM and TIL′E′ 26 nM ± 2 nM.
Binding analysis of TIL′E′ functionality. Solid-phase binding assays of recombinant FVIII to plasma-derived VWF (pdVWF; ▾), TIL′E′ expressed in E coli. (TIL′E′; ▪), and reduced and alkylated TIL′E′ (Red./alkyl. TIL′E′; ●). The obtained apparent dissociation constants are pdVWF 0.9 nM ± 0.2 nM and TIL′E′ 26 nM ± 2 nM.
NMR assignments, verification of disulfide-bond topology, and structure of the TIL′E′ domains
After establishing that the recombinantly produced TIL′E′ was both correctly folded and functional, the NMR resonances covering 78% of the TIL′E′ 1H, 13C, and 15N backbone nuclei were assigned using standard triple-resonance NMR experiments (Figure 2). The unassigned nuclei are predominantly located between residues 781 and 805 of the TIL′ domain (supplemental Figure 5), a consequence of resonance broadening due to micro-to-millisecond chemical exchange processes in this region of the protein (see below). The resonances of side-chain 1H and 13C nuclei were subsequently assigned using 3-dimensional carbon total correlated spectroscopy (CC-TOCSY)-based experiments to correlate the backbone resonances with those of the side chains,28-30 giving a total assignment of 89% of the TIL′E′ nuclei. 15N- and 13C-edited NOE spectroscopy experiments were used to obtain 954 unique interresidue distance restraints (supplemental Table 1; supplemental Figure 6) for structure determination. In addition, 108 backbone dihedral-angle restraints (supplemental Table 2) were derived from the TIL′E′ chemical shifts,31 and 361 vector-orientation restraints were collected by measurement of multiple RDCs in 2 alignment media (supplemental Table 3).32
The disulfide-bonding pattern shown in Figure 1 was verified by cross-disulfide NOEs corresponding to Cysi Hα/β-Cysj Hα/β, where Cysi-Cysj represents a predicted disulfide bond between the ith and jth cysteine residues in the domain sequence (supplemental Figure 7). This permitted the unambiguous assignment of 4 disulfide bridges, including Cys792-Cys827 in TIL′ and Cys829-Cys851, Cys849-Cys858, and Cys846-Cys863 in E′ that had been predicted9 but not previously biochemically assigned (Figure 1 and supplemental Figure 7). The overall disulfide-bonding topology was further confirmed by exclusion of the corresponding constraints from preliminary structure calculations. With the exception of the Cys788-Cys799 linkage and to some extent the Cys776-Cys804 linkage in the TIL′ domain, which were poorly defined due to limited NOE density, all predicted cysteine pairs came into close proximity. Therefore, all disulfide bonds were included as constraints in the structure calculation, resulting in the solution structure of VWF TIL′E′ shown in Figure 4.
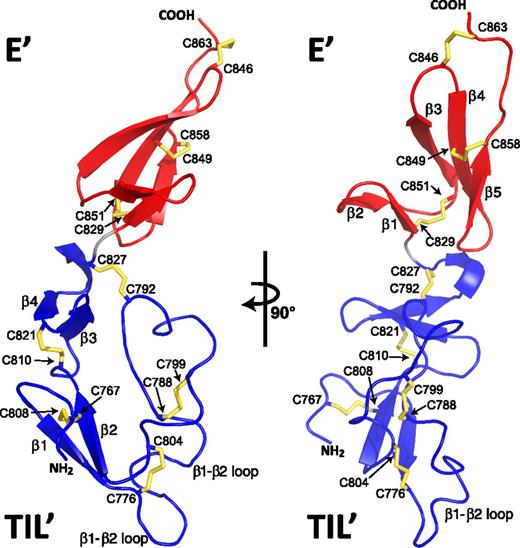
High-resolution solution structure of VWF TIL′E′. Ribbon diagram of the lowest-energy solution structure of TIL′E′ of VWF; disulfide bonds are colored yellow. The TIL′ domain (bottom) is shown in blue, and the E′ domain (top) is shown in red. The view of the structure shown to the right is a 90° rotation of the left view. β strands are numbered according to the order in which they occur in the primary sequence of the individual domains (Figure 1). Cysteine linkages are annotated with their residue numbers; a correlation with cysteine numbering within each of the 2 domains can be found in Figure 1.
High-resolution solution structure of VWF TIL′E′. Ribbon diagram of the lowest-energy solution structure of TIL′E′ of VWF; disulfide bonds are colored yellow. The TIL′ domain (bottom) is shown in blue, and the E′ domain (top) is shown in red. The view of the structure shown to the right is a 90° rotation of the left view. β strands are numbered according to the order in which they occur in the primary sequence of the individual domains (Figure 1). Cysteine linkages are annotated with their residue numbers; a correlation with cysteine numbering within each of the 2 domains can be found in Figure 1.
TIL′ has a long dominating loop, whereas E′ is all β sheet
The global fold of TIL′E′ shown in Figure 4 is crescent shaped, as has been suggested from electron microscopy imaging.5 The structure clearly shows that TIL′ and E′ form independently folding domains, confirming the previous domain assignment separating D′ into the 2 distinct domains.9 Salient features of domain TIL′ (residues 766-827) are the limited regions of secondary structure (∼70% loop; 26% β strand) and lack of a significant hydrophobic core, such that 5 conserved disulfide bonds constitute the major stabilizing force constraining the fold (Figure 4). TIL′ is formed of 2 short β sheets, β1:β2 (residues 772-775:806-809) and β3:β4 (residues 814-817:820-823), that form a scaffold bearing a long 30-residue loop, the β1-to-β2 loop. This loop encompasses an 8-residue insertion between the second and third conserved TIL′ cysteine residues as compared with structural homologs of TIL′ (supplemental Figure 1). The second antiparallel β sheet, formed of strands β3 and β4, is connected by a reverse turn forming a small-hairpin structure. There is a single turn of a 310 helix (residues 824-826) at the C terminus of TIL′. TIL′ belongs to the Ascaris protease inhibitor family, defined by a 10-cysteine signature and a conserved protein fold,33 with the closest structural homolog being a chymotrypsin inhibitor domain (Protein Data Bank accession number 1CCV) with a Z score of 3.6, as calculated by the DALI server.34 In contrast, Figure 4 shows that the greater proportion of E′ is formed of a triple-stranded antiparallel β sheet formed of strands β3 (residues 839-844), β4 (residues 847-852), and β5 (residues 855-858). Additionally, the N terminus contains a short double-stranded β1:β2 sheet (residues 829-831:834-836). The relative positions of the 2 β sheets are restrained by the E′ Cys829-Cys851 disulfide bond. The 6-cysteine E′ domain was previously suggested to be a member of the fibronectin I–like superfamily of proteins35 and thus similar to the von Willebrand factor type C (VWC) module. Supplemental Figure 8 shows an overlay of the structure of the E′ domain solved here by NMR spectroscopy and the first subdomain of a full VWC module. The 2 domains shown are structurally highly similar with a backbone root-mean-square deviation (RMSD) of 1.3 Å, thus highlighting the structural similarities between the E′ domain and the VWC module.
The family of the 10 lowest-energy structures of TIL′E′ is well defined, with an average backbone pairwise RMSD of 1.12 Å, and corresponding RMSDs for the TIL′ and E′ domains in isolation of 1.37 Å and 0.34 Å, respectively (supplemental Figure 9). The backbone RMSD of the TIL′ domain excluding the long β1-to-β2 loop is 0.81 Å, indicating that this loop dominates the structural heterogeneity of TIL′ and alluding to its intrinsic dynamic character (supplemental Figure 10). The 10 lowest-energy structures have 73.1% of residues in the most-favored regions in the Ramachandran plot, 22.3% in the additionally allowed regions, 3.4% in the generously allowed regions, and 1.2% in disallowed regions.
TIL′ is strikingly dynamic on at least 2 timescales
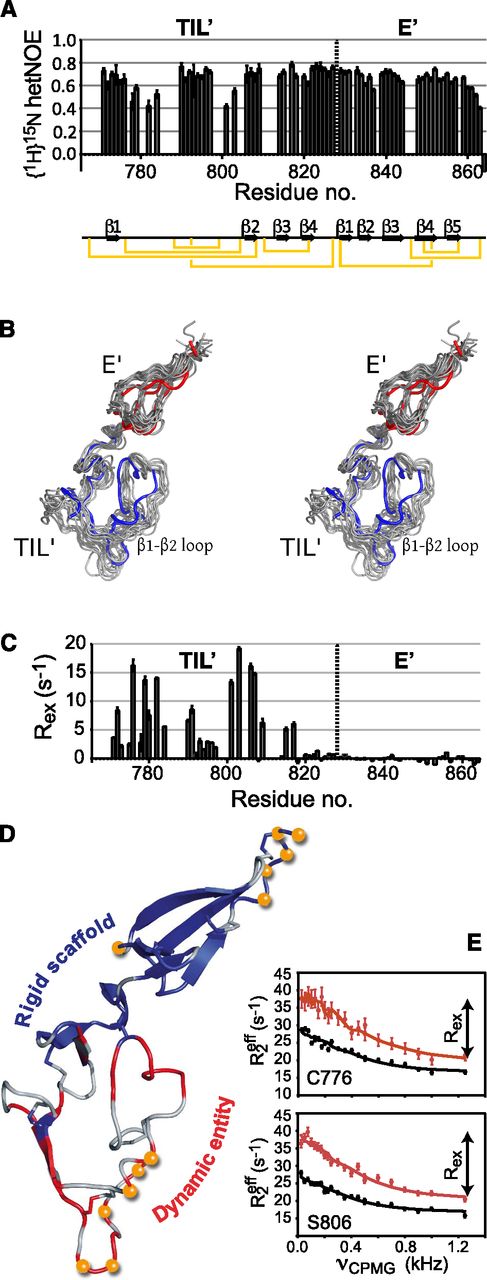
The conformational sampling of a protein is often essential to its biological activity,36 and hence the dynamic characterization of TIL′E′ is of great importance for elucidating the mechanism by which FVIII is stabilized in plasma. The structural dynamics of TIL′E′ were quantified experimentally by 15N nuclear spin relaxation experiments.17 Analysis of these relaxation data revealed interesting motional trends; most notably, that there is a distinguishable mobility differential between the TIL′ and E′ domains. {1H}15N nuclear Overhauser effects (hetNOEs; Figure 5A), which report specifically on backbone fluctuations on a pico-to-nanosecond (ps-ns) timescale, clearly show that, with the exception of the very C-terminal residues, the E′ domain is rigid with a uniform distribution of hetNOE values around 0.7. In contrast, several residues in the β1-to-β2 loop of the TIL′ domain show significantly lower NOE values, indicating a comparatively higher degree of flexibility. The amplitudes of the local ps-ns motions were quantified as squared order parameters,18 S2, according to the model-free analysis of the 15N spin relaxation rates (supplemental Table 4), and confirmed the relative flexibility of the TIL′ β1-to-β2 loop on this timescale.37 The analysis of the 15N relaxation rates includes a determination of the diffusion tensor, D. Of special interest here is the ratio of the parallel and the perpendicular components of the axial diffusion tensor, D||/D⊥, because this ratio reports on the shape of the molecule. Firstly, the ratios obtained separately for the 2 domains are very similar (D||,TIL′/D⊥,TIL′ = 1.82 ± 0.05 and D||,E′/D⊥,E′ = 1.9 ± 0.1), which indicates that the TIL′ and E′ domains tumble as a single entity with only limited interdomain motion. Secondly, the relatively large values of D||/D⊥ confirm that the overall structure is a highly prolate spheroid, in agreement with the calculated structures (Figure 4).
Pico-to-microsecond and millisecond structural dynamics of TIL′E′. (A) Backbone TIL′E′ {1H}15N heteronuclear NOEs that report on motions on the subnanosecond timescale. Low values indicate flexible, unstructured regions, whereas stabilizing structural elements give rise to higher hetNOEs. Secondary-structure elements of TIL′E′ are depicted below the plot. (B) Simultaneous stereo representation of the structure and pico-to-microsecond dynamics (gray) overlaid with the lowest-energy structure from the single-structure calculations (TIL′, blue; E′, red). The stereo mode is wall-eye. (C) Nitrogen backbone millisecond chemical exchange contributions,  , obtained at 700 MHz. Significant exchange contributions, Rex, indicate dynamics on a microsecond to millisecond timescale (conformational exchange). Backbone millisecond dynamics are almost entirely confined to the TIL′ domain, with the E′ domain comparatively rigid on this timescale. (D) Combined dynamics of TIL′E′; orange balls represent residues with {1H}15N NOE <0.6, whereas the backbone ribbon is colored red (blue) for residues whose Rex is greater (less) than 1.5 s−1. The backbone for residues where no Rex is available is colored gray. (E) Two representative dispersion profiles of residues with millisecond structural dynamics, Cys776 and Ser806. Points (vertical bars) represent measured Rex values (uncertainties), and the full-drawn line is a least-squares fit of a 2-site conformational exchange model to the data shown (red. 700 MHz; black, 500 MHz).
, obtained at 700 MHz. Significant exchange contributions, Rex, indicate dynamics on a microsecond to millisecond timescale (conformational exchange). Backbone millisecond dynamics are almost entirely confined to the TIL′ domain, with the E′ domain comparatively rigid on this timescale. (D) Combined dynamics of TIL′E′; orange balls represent residues with {1H}15N NOE <0.6, whereas the backbone ribbon is colored red (blue) for residues whose Rex is greater (less) than 1.5 s−1. The backbone for residues where no Rex is available is colored gray. (E) Two representative dispersion profiles of residues with millisecond structural dynamics, Cys776 and Ser806. Points (vertical bars) represent measured Rex values (uncertainties), and the full-drawn line is a least-squares fit of a 2-site conformational exchange model to the data shown (red. 700 MHz; black, 500 MHz).
Pico-to-microsecond and millisecond structural dynamics of TIL′E′. (A) Backbone TIL′E′ {1H}15N heteronuclear NOEs that report on motions on the subnanosecond timescale. Low values indicate flexible, unstructured regions, whereas stabilizing structural elements give rise to higher hetNOEs. Secondary-structure elements of TIL′E′ are depicted below the plot. (B) Simultaneous stereo representation of the structure and pico-to-microsecond dynamics (gray) overlaid with the lowest-energy structure from the single-structure calculations (TIL′, blue; E′, red). The stereo mode is wall-eye. (C) Nitrogen backbone millisecond chemical exchange contributions,  , obtained at 700 MHz. Significant exchange contributions, Rex, indicate dynamics on a microsecond to millisecond timescale (conformational exchange). Backbone millisecond dynamics are almost entirely confined to the TIL′ domain, with the E′ domain comparatively rigid on this timescale. (D) Combined dynamics of TIL′E′; orange balls represent residues with {1H}15N NOE <0.6, whereas the backbone ribbon is colored red (blue) for residues whose Rex is greater (less) than 1.5 s−1. The backbone for residues where no Rex is available is colored gray. (E) Two representative dispersion profiles of residues with millisecond structural dynamics, Cys776 and Ser806. Points (vertical bars) represent measured Rex values (uncertainties), and the full-drawn line is a least-squares fit of a 2-site conformational exchange model to the data shown (red. 700 MHz; black, 500 MHz).
, obtained at 700 MHz. Significant exchange contributions, Rex, indicate dynamics on a microsecond to millisecond timescale (conformational exchange). Backbone millisecond dynamics are almost entirely confined to the TIL′ domain, with the E′ domain comparatively rigid on this timescale. (D) Combined dynamics of TIL′E′; orange balls represent residues with {1H}15N NOE <0.6, whereas the backbone ribbon is colored red (blue) for residues whose Rex is greater (less) than 1.5 s−1. The backbone for residues where no Rex is available is colored gray. (E) Two representative dispersion profiles of residues with millisecond structural dynamics, Cys776 and Ser806. Points (vertical bars) represent measured Rex values (uncertainties), and the full-drawn line is a least-squares fit of a 2-site conformational exchange model to the data shown (red. 700 MHz; black, 500 MHz).
Backbone dynamics on the broader pico-to-microsecond (ps-μs) timescale were investigated, because ligand binding and dissociation often takes place in this time regime.36,38 The amplitudes of motions over the ps-μs timescale were estimated from backbone chemical shifts31 and subsequently included in structure calculations,21,39 together with the restraints listed above, to determine an ensemble of protein conformations that represent, simultaneously, the TIL′E′ structure and its associated dynamics. Accordingly, this structural ensemble represents a more faithful atomistic description of the conformational variability of TIL′E′ on a ps-μs timescale. The ensemble representation of TIL′E′ was systematically cross-validated by a combination of RDCs and side-chain 13C methyl chemical shifts40-43 to give an optimal ensemble size of 10 replicas (supplemental Figure 11). The ensemble representation is shown in Figure 5B and shows that the β1-to-β2 loop is significantly more flexible than the remainder of TIL′E′.
Although the hetNOEs (Figure 5A) and chemical shifts report on ps-ns and ps-μs dynamics, respectively, structural dynamics on the millisecond timescale can be specifically probed using CPMG relaxation dispersion experiments.19,44 The time constants of many biochemical processes, such as ligand binding and dissociation,36 are on the millisecond timescale, and CPMG experiments were therefore performed on the TIL′E′ domains to quantify such dynamics (Figure 5C). A first inspection of the CPMG data reveals localization of slower motions to the domain TIL′, particularly in the β1-to-β2 loop (Figure 5C), confirming the existence of a sparsely populated (excited) state with a lifetime in the millisecond range, whereas the E′ domain remains rigid on this timescale. A detailed analysis of the CPMG relaxation dispersions reveals that the ground state of TIL′E′ exchanges with an excited state; the lifetime of the excited state is approximately 0.5 ms and has a population of 3.5% (supplemental Methods and materials). Excited states often have significant functional roles in molecular recognition and ligand binding, and the existence of the TIL′E′ excited state is likely to be functionally relevant for the binding of VWF to FVIII.
A summary of the TIL′E′ dynamics is shown in Figure 5D, from which it is evident that the TIL′ β1-to-β2 loop is strikingly dynamic on at least 2 timescales, whereas the remainder of TIL′E′ provides a rigid scaffold supporting this conformationally dynamic entity.
Discussion
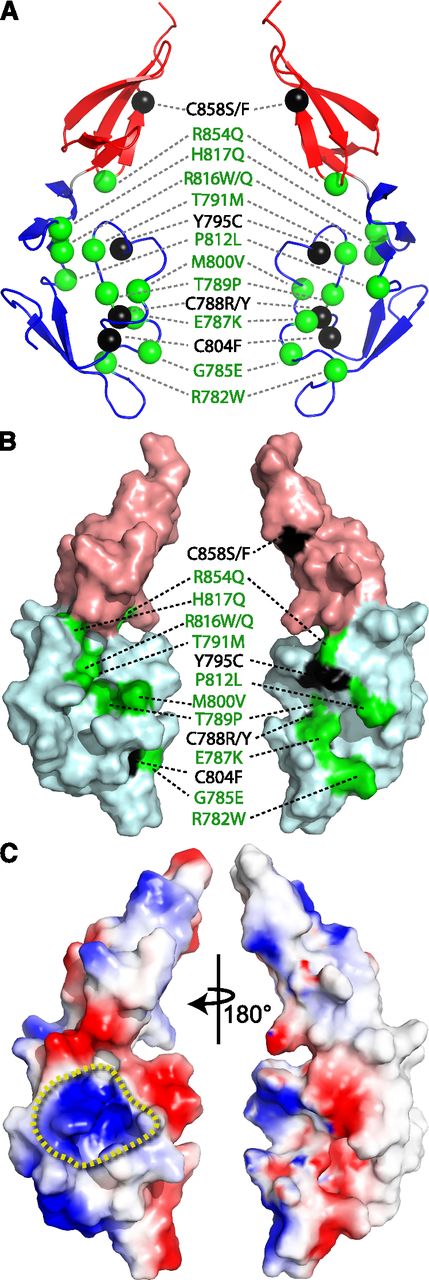
The solution structure and dynamic characterization of TIL′E′ allow an unprecedented interpretation of the likely functional consequence of type 2N VWD missense mutations. The greater proportion of the known type 2N missense mutations are distributed in the vicinity of the dynamic TIL′ β1-to-β2 loop (Figure 6A). Type 2N mutations that involve a cysteine residue (C788R/Y, Y795C, and C804F in TIL′; C858S/F in E′) are associated with aberrant multimerization, poor secretion, and reduced FVIII binding,45 indicating that correct disulfide-bond formation is critical to the folding and function of VWF (Figure 3). Of the type 2N mutations, 3 account for the majority of cases reported (T791M, R816W, and R854Q),10 of which R854Q occurs with polymorphic frequency,4 and all of which are in immediate proximity to the β1-to-β2 loop. However, the most accurate reporters of the FVIII binding site are those mutations that are associated with a more severe type 2N phenotype; that is, E787K, T791M, and R816W.10 Two of these mutations, T791M and R816W, together with T789P, M800V, R816Q, and H817Q, are clustered around a region of positive charge density on TIL′ (Figure 6B-C). Two regions on the FVIII light chain have been implicated in binding VWF, that is, the N-terminal acidic a3 domain and the C-terminal C2 domain. Although these 2 regions are thought to act synergistically in binding VWF, studies show that a3 in isolation binds ∼8 times more strongly to VWF than the C2 domain, with affinities of ∼72 nM and 564 nM,46 respectively, and the acidic a3 domain is critical to high-affinity binding to VWF.46 We therefore hypothesize that the positively charged and dynamic region on TIL′E′ is the major binding site to the negatively charged FVIII a3 domain. Thus, the deleterious effect of the R816W mutant is likely due to the introduction of a large and hydrophobic side chain and/or loss of positive charge (Table 1 47 ). Figure 6 shows that residues R782, G785, and E787 are on the opposite side of the structure yet in close proximity to the β1-to-β2 loop (Figure 6 and supplemental Figure 12), which suggests that the severe type 2N mutation E787K, and mutations R782W and G785E, lead to deficiencies in FVIII binding through a perturbation of the β1-to-β2 loop, the upper portion of which borders the predicted binding region.
Structural interpretation of type 2N VWD mutations. (A) Ribbon diagram showing the position of the reported TIL′E′ type 2N missense mutations. Noncysteine mutations are shown with green spheres, and those involving cysteine residues are colored black. The TIL′ domain is colored blue, and the E′ domain colored red. A wall-eye stereo view is shown in supplemental Figure 12. (B) Surface representation of TIL′E′ with the type 2N noncysteine mutations colored green, and those involving cysteine residues colored black. The surface of the TIL′ domain is shown with a light-blue color, and the surface of the E′ domain is shown with a light-red color. (C) Electrostatic surface potential of TIL′E′, with blue (red) indicating positive (negative) charge. The view on the left side shows the region of concentrated positive charge density, circumscribed by a yellow dashed line, which is the putative binding site for the a3 domain of the FVIII light chain. The views on the right side of all panels are a 180° rotation of the views shown on the left.
Structural interpretation of type 2N VWD mutations. (A) Ribbon diagram showing the position of the reported TIL′E′ type 2N missense mutations. Noncysteine mutations are shown with green spheres, and those involving cysteine residues are colored black. The TIL′ domain is colored blue, and the E′ domain colored red. A wall-eye stereo view is shown in supplemental Figure 12. (B) Surface representation of TIL′E′ with the type 2N noncysteine mutations colored green, and those involving cysteine residues colored black. The surface of the TIL′ domain is shown with a light-blue color, and the surface of the E′ domain is shown with a light-red color. (C) Electrostatic surface potential of TIL′E′, with blue (red) indicating positive (negative) charge. The view on the left side shows the region of concentrated positive charge density, circumscribed by a yellow dashed line, which is the putative binding site for the a3 domain of the FVIII light chain. The views on the right side of all panels are a 180° rotation of the views shown on the left.
Type 2N missense mutations in domains TIL′ and E′ of VWF
| Domain . | Mutation . | Δ Volume (Å3)* . | Position . | Predicted effect . |
|---|---|---|---|---|
| TIL′ | R782W | +41.1 | β1-β2 loop | Loss +ve, polar-to-hydrophobic, structural perturbation |
| G785E | +88.1 | β1-β2 loop | Introduce −ve charge, ↑volume | |
| E787K | +8.9 | β1-β2 loop | Introduce +ve in region of −ve charge, highly deleterious | |
| C788R | +94.7 | β1-β2 loop | Introduction of -SH, introduce +ve, ↑volume | |
| C788Y | +108.4 | β1-β2 loop | Introduction of -SH, ↑volume | |
| T789P | −3.6 | β1-β2 loop | Perturbation of putative a3 binding region | |
| T791M | +40.9 | β1-β2 loop | Perturbation of putative a3 binding region | |
| Y795C | −96.4 | β1-β2 loop | Exposed -SH | |
| M800V | −28.5 | β1-β2 loop | Border of putative a3 binding region | |
| C804F | +85.4 | β1-β2 loop | Introduction of -SH | |
| P812L | +41.0 | β2-β3 loop | Perturbation of putative a3 binding region | |
| R816W | +42.1 | β3 | Loss of conserved +ve in putative a3 binding region | |
| R816Q | −40.7 | β3 | Loss of conserved +ve in putative a3 binding region | |
| H817Q | −7.8 | β3 | Loss of conserved +ve in putative a3 binding region | |
| E′ | R854W | +42.1 | β4-β5 loop | Loss +ve, polar-to-hydrophobic |
| R854Q | −40.7 | β4-β5 loop | Loss +ve | |
| C858F | +97.4 | β5 | Introduction of -SH | |
| C858S | −0.4 | β5 | Introduction of -SH |
| Domain . | Mutation . | Δ Volume (Å3)* . | Position . | Predicted effect . |
|---|---|---|---|---|
| TIL′ | R782W | +41.1 | β1-β2 loop | Loss +ve, polar-to-hydrophobic, structural perturbation |
| G785E | +88.1 | β1-β2 loop | Introduce −ve charge, ↑volume | |
| E787K | +8.9 | β1-β2 loop | Introduce +ve in region of −ve charge, highly deleterious | |
| C788R | +94.7 | β1-β2 loop | Introduction of -SH, introduce +ve, ↑volume | |
| C788Y | +108.4 | β1-β2 loop | Introduction of -SH, ↑volume | |
| T789P | −3.6 | β1-β2 loop | Perturbation of putative a3 binding region | |
| T791M | +40.9 | β1-β2 loop | Perturbation of putative a3 binding region | |
| Y795C | −96.4 | β1-β2 loop | Exposed -SH | |
| M800V | −28.5 | β1-β2 loop | Border of putative a3 binding region | |
| C804F | +85.4 | β1-β2 loop | Introduction of -SH | |
| P812L | +41.0 | β2-β3 loop | Perturbation of putative a3 binding region | |
| R816W | +42.1 | β3 | Loss of conserved +ve in putative a3 binding region | |
| R816Q | −40.7 | β3 | Loss of conserved +ve in putative a3 binding region | |
| H817Q | −7.8 | β3 | Loss of conserved +ve in putative a3 binding region | |
| E′ | R854W | +42.1 | β4-β5 loop | Loss +ve, polar-to-hydrophobic |
| R854Q | −40.7 | β4-β5 loop | Loss +ve | |
| C858F | +97.4 | β5 | Introduction of -SH | |
| C858S | −0.4 | β5 | Introduction of -SH |
Changes in residue volumes are based on Pontius et al.47
In summary, the majority of the noncysteine type 2N mutations (Figure 6) are found in the vicinity of the flexible regions of TIL′, thereby localizing the FVIII binding site to this part of the VWF protein. Of the type 2N mutations in TIL′E′ involving charged residues, all but one result in loss of positive charge, supporting a predominantly electrostatic interaction between VWF and the FVIII light chain, which is in agreement with the fact that high ionic strength and low pH result in a much lower affinity of the VWF⋅FVIII complex.27,48 Hence, based on the foregoing analyses, we propose that E′ and the β sheets of TIL′ provide a rigid scaffold for the positively charged region on TIL′, which complements the charge distribution of the FVIII light chain to electrostatically guide VWF⋅FVIII complex formation.
The cysteine-rich N- and C-terminal flanking regions of VWF have been the subject of significant efforts aimed at characterizing the structure and plasticity of function of this unique protein. Superposition of the VWD type 2N missense mutations on the structure of TIL′E′, together with the characterization of its structural dynamics, reveals the malleable FVIII binding surface of VWF and represents an important step toward a detailed molecular understanding of these functionally important VWF domains for which atomic-resolution structures have hitherto been unavailable.
Presented as an oral communication at the XXIV congress of the International Society on Thrombosis and Haemostasis, Amsterdam, the Netherlands, July 1, 2013.
The atomic coordinates have been deposited in the Protein Data Bank (http://www.pdb.org) with the accession numbers 2mhp (single structure) and 2mhq (ensemble), and the chemical-shift assignments have been deposited in the Biological Magnetic Resonance Data Bank (http://www.bmrb.wisc.edu) with the code 19646.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr P. J. Lenting for providing the mAb-418, A. Riddell for helpful discussions and for providing the recombinant FVIII, and Prof S. K. Srai, Dr K. Gomez, and Prof G. Waksman for helpful discussions. The authors acknowledge the Medical Research Council and the National Institute for Medical Research for access to the 800 MHz spectrometer for exploratory characterizations of TIL′E′.
This work was supported by the International Journal of Experimental Pathology and the Comprehensive Biomedical Research Centre (studentship to N.S.), the Rosetrees Trust, Novo Nordisk A/S, and the Biotechnology and Biological Sciences Research Council (BBSRC).
Authorship
Contribution: N.S., E.G.D.T., and D.F.H. designed the research; N.S., J.K., and D.F.H. performed and analyzed the NMR experiments and NMR data; N.S. and D.F.H. performed structure calculations; N.S. and L.D.C. cloned, expressed, and purified isotope-labeled TIL′E′; N.S. and L.D.C. designed and performed dot-blot analyses; K.T. performed MS studies; T.A.J.M. performed solid-phase binding assays; and all authors discussed the results and contributed to writing the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: D. Flemming Hansen, Institute of Structural and Molecular Biology, University College London, Gower St, London, WC1E 6BT, United Kingdom; e-mail: d.hansen@ucl.ac.uk; or Edward G. D. Tuddenham, The Katharine Dormandy Haemophilia Centre and Thrombosis Unit, Royal Free Hospital NHS Trust, Pond St, London, NW3 2QG, United Kingdom; e-mail: e.tuddenham@ucl.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal