Key Points
A population-specific genetic variant involved in folate homeostasis is associated with lower warfarin dose in African Americans.
Abstract
The anticoagulant warfarin has >30 million prescriptions per year in the United States. Doses can vary 20-fold between patients, and incorrect dosing can result in serious adverse events. Variation in warfarin pharmacokinetic and pharmacodynamic genes, such as CYP2C9 and VKORC1, do not fully explain the dose variability in African Americans. To identify additional genetic contributors to warfarin dose, we exome sequenced 103 African Americans on stable doses of warfarin at extremes (≤35 and ≥49 mg/week). We found an association between lower warfarin dose and a population-specific regulatory variant, rs7856096 (P = 1.82 × 10−8, minor allele frequency = 20.4%), in the folate homeostasis gene folylpolyglutamate synthase (FPGS). We replicated this association in an independent cohort of 372 African American subjects whose stable warfarin doses represented the full dosing spectrum (P = .046). In a combined cohort, adding rs7856096 to the International Warfarin Pharmacogenetic Consortium pharmacogenetic dosing algorithm resulted in a 5.8 mg/week (P = 3.93 × 10−5) decrease in warfarin dose for each allele carried. The variant overlaps functional elements and was associated (P = .01) with FPGS gene expression in lymphoblastoid cell lines derived from combined HapMap African populations (N = 326). Our results provide the first evidence linking genetic variation in folate homeostasis to warfarin response.
Introduction
With >33 million prescriptions in 2011, warfarin is the most commonly used anticoagulant for preventing thromboembolic events.1 Warfarin has a 20-fold interindividual dose variability and a narrow therapeutic index, and it is responsible for one-third of adverse drug event hospitalizations in older Americans.2 Alternatives to warfarin, such as direct thrombin inhibitors and factor Xa inhibitors, are now available. However, these are more expensive, irreversible, and may cause a higher rate of acute coronary events compared with warfarin.3,4 Thus, warfarin remains a mainstay of anticoagulant therapy, and better methods of dosing warfarin will lead to fewer adverse events.
In addition to clinical modifiers, genetic polymorphisms are also known to affect an individual’s stable therapeutic warfarin dose.5 These polymorphisms modulate warfarin’s pharmocodynamic or pharmacokinetic pathways and include variants in genes coding for the warfarin’s target, the vitamin K epoxide reductase complex, subunit 1 (VKORC1), and warfarin’s main metabolizer, cytochrome p450 family 2, subfamily C, polypeptide 9 (CYP2C9).6,7 The Food and Drug Administration approved the addition of dosing recommendations based on genotype to warfarin labeling.8 In 2012, Anderson et al demonstrated in a clinical effectiveness trial that using pharmacogenetics to predict therapeutic warfarin dose is superior than the standard of care in establishing and maintaining therapeutic anticoagulation, as measured by international normalized ratio, and may reduce the risk for adverse events.8
In 2013, as part of the Clarification of Optimal Anticoagulation through Genetics (COAG) trial, Kimmel et al conducted a double-blind randomized controlled trial that showed no difference in time in therapeutic range over 4 weeks when dosing warfarin with a pharmacogenetic algorithm vs a clinical algorithm.9,10 The implications of these findings are unclear because clinical algorithms are not the current standard of care.10 In parallel, Pirmohamed et al conducted a single-blind randomized controlled trial and showed the superiority of using a modified version of the International Warfarin Pharmacogenetics Consortium (IWPC) pharmacogenetic equation over the current standard of care in the United Kingdom and Sweden.11 Although both of these studies focused on European populations, Kimmel et al included 275 African American subjects and concluded that the tested variants performed poorly, leading to a significant overprediction of dose.9 Importantly, VKORC1 −1639 G/A, CYP2C9*2, and CYP2C9*3, the only variants studied by Kimmel et al, are less prevalent in African Americans, who have some of the greatest known dose variability.12-14 Thus, this study highlighted the need for population-specific data in warfarin pharmacogenetics. Variants relevant to warfarin dose with greater frequency in African Americans have been discovered through candidate gene studies, targeted sequencing of the warfarin pharmacodynamics and pharmacokinetic pathways, and a genome-wide association study.15,16 For instance, a variant in a VKORC1 regulator, calumenin (CALU), leads to higher predicted warfarin doses in African Americans, whereas the CYP2C9*8 variant leads to lower doses.17-19 Additionally, a recent genome-wide association study showed a variant in the CYP2C cluster associates with lower warfarin dose in African Americans.15 However, these genetic factors and all the known clinical variables explain only 29% to 41% of the variability in warfarin dose in this population compared with >50% in European descent populations.5,19
Therefore, identifying additional genetic factors that affect warfarin dose in populations of African descent is critical for extending the benefits of genotype guidance to these populations. To this end, we used an extreme phenotype strategy paired with exome sequencing to find novel variants that influence warfarin dosing among African Americans.20 We report a novel association between a genetic variant affecting folate homeostasis and lower warfarin dose requirement among African Americans.
Methods
Discovery cohort
Selection.
The discovery cohort was selected using the dose distribution of 360 African Americans on warfarin and selecting individuals who were at least ∼0.25 standard deviations away from the mean: 44.5 mg/week (supplemental Figure 1).15 The cutoffs were 35 mg/week for the low-dose cohort and 49 mg/week for the high-dose cohort. One hundred six individuals met these criteria. Samples were collected from sites at the University of Chicago, University of Illinois at Chicago, and University of Florida. All patients consented using an Institutional Review Board–approved process at each respective collection site in accordance with the Declaration of Helsinki.
Sequencing, mapping, and variant calling.
Exome capture and sequencing were performed using 2.5 to 3 µg of DNA (concentration ≥ 200 ng/µL) with Illumina’s TruSeq v1 and sequenced using the Illumina GAIIx platform. Reads were mapped using BWA 0.6.1 (with the -q 20 flag). Duplicate reads were marked with Picard 1.56, and local realignment around potential indels (including the Single Nucleotide Polymorphysm database [dbSNP] 132) was performed with the Genome Analysis Toolkit (GATK; 1.3-21-gcb284ee). GATK was used to recalibrate quality scores with dbSNP 132 as a training set and to calculate depth of coverage.
Variant calling.
SNPs were called with GATK's UnifiedGenotyper only at sites covered by the exome sequencing platform and the 50 bp surrounding these sites. Quality scores were calibrated with Variant Quality Score Recalibration using the default settings. VCFtools was used to calculate the missing data rate, heterozygosity, and depth per site, which was redundant with depth from mapping. Indels were called using GATK’s Unified Genotyper, and quality scores were calibrated with Variant Quality Score Recalibration at default settings. With quality score filtering, we called ∼500 000 SNPs.
Quality control.
We submitted 1 sample from a subject of European descent who had previously undergone exome sequencing at 30× coverage by the same Illumina platform.21 Moreover, 86 samples had been previously genotyped on the Illumina 610 Quad BeadChip (Illumina, San Diego, CA). Concordance between the previously exome sequenced sample of European descent and the high-quality exome calls was 99.8%.
As an additional quality control step, we confirmed that identity by descent (IBD) analysis showed that 85 of 86 exome-sequenced samples shared 100% IBD with their genotyped data. One sample had a concordance of 79%; this sample had the lowest coverage (mean coverage ∼18×) and was removed from subsequent analyses. The mean depth of coverage of the remaining samples was about 68×. Finally, IBD revealed that 3 samples were unexpectedly related; this relationship was confirmed by consulting the clinical charts. For the final analysis, we removed 2 of these samples so that the samples used for analysis were completely unrelated. Samples had previously been shown to meet the criteria for African-American descent.15
In addition to quality score filtering, we filtered out SNPs with a minor allele frequency <0.05 and loci missing calls in >20 individuals (∼20% missingness). We only tested a total of 155 186 SNPs.
Clinical data.
Demographic and clinical data were collected as previously described.14,15 Collected clinical covariates included gender, age, height, weight, concurrent use of aspirin, amiodarone, enzyme inducers (rifampin, phenytoin, and carbamazepine), indication for warfarin, and stable dose of warfarin. One individual was missing height, which was estimated by using the sex-matched mean. One individual was missing dose indication, and this was set to the default of no venous thromboembolism (VTE).
Analysis.
Single variant analysis.
A total of 103 unrelated samples met the above quality control requirements. Genotyped covariates were VKORC1 −1639G>A (rs9923231), CYP2C9*2 (rs1799853), CYP2C9*3 (rs1057910), CYP2C9*5 (rs28371686), CYP2C9*8 (rs7900194), and CYP2C9*11 (rs28371685). The VKORC1 −1639G>A genotype was modeled as an additive covariate, whereas the CYP2C9 haplotype was modeled as a composite additive covariate.
To account for population stratification, we used principal component analysis, which was calculated using the Nonlinear Iterative Partial Least Squares algorithm in the Principal Component Analysis module python package (version 1.1.02).22
We selected which clinical covariates and previously identified genetic covariates to use in our model based on univariate association with log-transformed warfarin dose (supplemental Table 1, available on the Blood Web site) using the linear model (lm) function in the R statistical package (version 2.15.3). We modeled log-transformed dose on these significant covariates and on every SNP that passed the aforementioned quality control requirements using linear model function in the R statistical package (version 2.15.3). P values were adjusted using Bonferroni correction, with a significance cutoff of 3.22 × 10−7 based on the 155 186 SNPs tested. Linkage disequilibrium (LD) between top SNPs was evaluated using PLINK (version 1.07).23
Gene-based analysis.
Previous extreme-phenotype sequencing studies have performed rare variant burden tests across genes to identify genes of interest.20 We used this same test, the RTV1 test, to perform a gene-based evaluation of our exomes.24 We mapped variants within RefSeq exon boundaries and used an allele frequency cutoff frequency of <0.125. For 42 284 evaluated reference sequences, we used a Bonferroni cutoff of 1.18 × 10−6.
Separately, we also specifically evaluated the presence of rare, damaging mutations in known pharmacokinetic and pharmacodynamic genes involved in warfarin dosing: CYP2C9 and VKORC1. Known CYP2C9 variants had been genotyped previously.
Replication cohort
The replication cohort consisted of 479 African Americans on a stable dose of warfarin enrolled from the University of Chicago, University of Illinois at Chicago, University of Florida, and University of Alabama at Birmingham; these samples were independent from the original 360 samples used for the extreme-phenotype study design in the discovery cohort. All patients provided written, informed consent for study participation. In the replication cohort, samples were not selected for an extreme dose distribution (supplemental Figure 2).
Genotyping.
Samples from University of Chicago, University of Illinois, and the University of Florida were genotyped via pyrosequencing, as previously described.25 A list of primers can be found in the supplemental Table 2.7 Genotypes for rs7856096 in the University of Alabama at Birmingham cohort were obtained from Genome-Wide Association Study Illumina Human 1M Duo 3.0 genotype data (directly interrogated) as previously described.15
Quality control.
Because race was self-reported, we tested ancestry as described below. Samples from the University of Alabama at Birmingham were assessed for ancestry using principal component analysis, as previously described.15 Samples from the University of Chicago and University of Illinois were assessed for ancestry using ancestry informative markers using previously described methods.26,27 In the replication, 83 subjects were found to have a significantly lower percentage of African ancestry based on previously described cutoffs and were removed prior to analysis.26,27
Clinical data.
Available clinical data from the replication data set included gender, age, height, weight, amiodarone use, and VTE status. Missing height and weight were imputed using the gender-matched mean.
Analysis.
A total of 372 samples met the quality control and missingness criteria. Genetic covariates included VKORC1 −1639G>A coded as an additive genetic variant and CYP2C9 haplotype data as a composite score. The University of Alabama at Birmingham data included a CYP2C9 composite score based on variants in CYP2C9*2, CYP2C9*3, CYP2C9*5, CYP2C9*6, and CYP2C9*11. The University of Chicago and University of Illinois CYP2C9 composite scores included information about the presence of CYP2C9*2, CYP2C9*3, CYP2C9*6, CYP2C9*8, and CYP2C9*11. For the replication model, we selected the clinical covariates that were used in the discovery model that were available. There were no data available on aspirin use in the replication cohort, and SNP array data were not available on the entire data set for principal component analysis. Thus, the reduced model used on the replication cohort incorporated age, weight, amiodarone, VTE status, VKORC1 −1639G>A, and CYP2C9 composite score. Log2-transformed dose was modeled using these covariates and the top hit using the linear model function in the R statistical package (version 2.15.3). Because we tested only the 1 SNP that was significant in our discovery cohort, we did not correct for multiple hypothesis testing.
We ran this same reduced model on the discovery cohort to ensure that the reduced model did not affect the significance of our top hit.
Information added to the IWPC model
We then evaluated the contribution of the SNP to the IWPC pharmacogenetic algorithm. We calculated the predicted warfarin dose of each individual in both the discovery and replication cohorts using the IWPC dose equation.5 We treated CYP2C9*11 and CYP2C9*8 as equivalent to CYP2C9*2 and CYP2C9*5 as equivalent to CYP2C9*3 and CYP2C9*6 as equivalent to CYP2C9 unknown.5,15,16 The residual dose unexplained by the equation was defined as the predicted dose subtracted from the actual dose. The discovery cohort included information on rifampin, carbamazepine, and phenytoin use.
For the replication cohort, participants from the University of Alabama at Birmingham were not on enzyme inducers, and data on carbamazepine use were available from the University of Chicago and University of Illinois at Chicago replication cohorts.
The novel SNP was analyzed as an additive variant, and we computed the additional information contributed to this residual dose using the lm function in R (version 2.15.3).
Functional analysis
We tested whether rs7856096 overlaps experimentally predicted functional elements from the Encyclopedia of DNA Elements (ENCODE) and the National Institutes of Health Epigenomics Roadmap.28,29 We downloaded chromatin state segmentation data (generated by ENCODE/Broad using ChIP-seq data) derived from 9 human cell types from the University of California, Santa Cruz Genome Browser.30
We also analyzed lymphoblastoid cell line (LCL) microarray gene expression data in the 3 HapMap African populations: the Maasai from Kenyawa, Kenya (MKK, N = 135), the Luhya in Webuye, Kenya (LWK; N = 83), and the Yoruba in Ibadan, Nigeria (YRI; N = 108).31 In particular, we tested, assuming an additive model, the effect of each risk allele on the expression of FPGS using the lm function in R (version 2.15.4), with or without population indicator as a covariate.
Results
Discovery cohort: rs7856096 significantly associated with dose in the extreme phenotype study
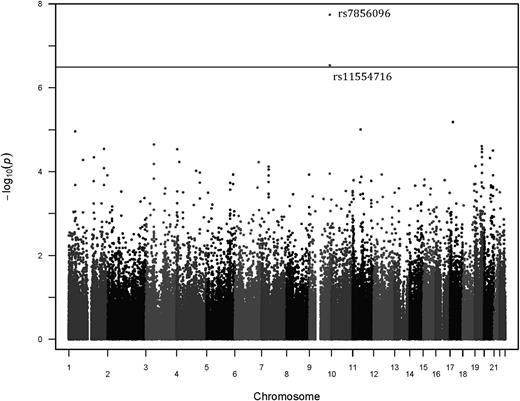
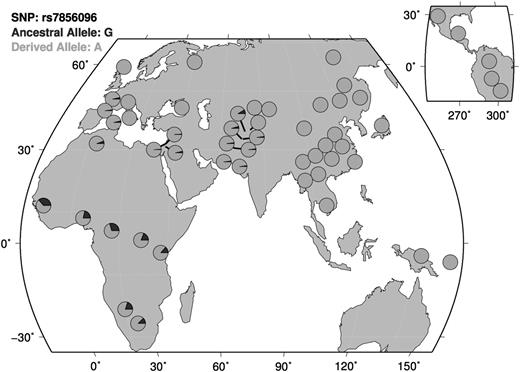
Our discovery cohort consisted of 103 unrelated African Americans on stable doses of warfarin. These subjects were selected for an extreme phenotype distribution. Population demographics and clinical data were collected as previously described and are summarized in Table 1.14 The allele frequency of VKORC1 −1639G>A in this cohort was 0.117. For CYP2C9 composite score, 15.5% of individuals had only 1 CYP2C9 and 6.8% had 2 CYP2C9 variants. Variables that significantly explained dose in a univariate model were age, weight, aspirin use, amiodarone use, warfarin indication, VTE status, VKORC1 −1639G>A, and composite CYP2C9 status (supplemental Table 1). The top 10 SNPs and P values from a linear model of log2-transformed warfarin dose are shown in Table 2. The coefficients for the covariates in this linear model can be found in supplemental Table 3. Only 1 SNP, rs7856096, was significantly associated with warfarin dose (uncorrected P value: 1.82 × 10−8; Figure 1). rs7856096 is upstream of exon 2 of the folylpolyglutamate synthase gene (FPGS), which encodes a mitochondrial enzyme involved in folate sequestration. This SNP was in LD with the second most significant SNP, rs11554716, with an R2 value of 0.85. An LD plot using the HapMap Phase III ASW data can be found in refs. 32 and 33 . Figure 2 shows a QQ-plot of expected vs observed P values. The risk allele, which was the ancestral allele, is most prevalent on the African continent (Figure 3) and has a frequency of 0.204 in our discovery population.32,34,35
Clinical demographics of the discovery cohort and replication cohort
| Covariate . | Discovery mean/% (all) (n = 103) . | Discovery mean/% (>49 mg/week) (n = 58) . | Discovery mean (<35 mg/week) (n = 45) . | Replication cohort mean/% . |
|---|---|---|---|---|
| Dose | 49.16 mg/wk | 66.38 mg/wk | 26.97 mg/wk | 43.80 mg/wk |
| Gender | 68.00% female | 67.2% female | 68.89% female | 61.39% female |
| Age | 56.75 y | 50.38 y | 64.96 y | 57.58 y |
| Height | 168.20 cm | 168.83 cm | 167.40 cm | 169.60 cm |
| Weight | 92.37 kg | 96.43 kg | 87.13 kg | 92.05 kg |
| Aspirin | 26.21% | 12.07% | 44.44% | — |
| Amiodarone | 0.97% | 0% | 2.22% | 4.30% |
| Phenytoin | 0.97% | 0% | 2.22% | — |
| Carbamazepine | 1.94% | 3.45% | 0% | — |
| Rifampin | 2.91% | 3.45% | 2.22% | — |
| Indication (VTE) | 64.71% | 82.75% | 40.91% | 47.04% |
| Covariate . | Discovery mean/% (all) (n = 103) . | Discovery mean/% (>49 mg/week) (n = 58) . | Discovery mean (<35 mg/week) (n = 45) . | Replication cohort mean/% . |
|---|---|---|---|---|
| Dose | 49.16 mg/wk | 66.38 mg/wk | 26.97 mg/wk | 43.80 mg/wk |
| Gender | 68.00% female | 67.2% female | 68.89% female | 61.39% female |
| Age | 56.75 y | 50.38 y | 64.96 y | 57.58 y |
| Height | 168.20 cm | 168.83 cm | 167.40 cm | 169.60 cm |
| Weight | 92.37 kg | 96.43 kg | 87.13 kg | 92.05 kg |
| Aspirin | 26.21% | 12.07% | 44.44% | — |
| Amiodarone | 0.97% | 0% | 2.22% | 4.30% |
| Phenytoin | 0.97% | 0% | 2.22% | — |
| Carbamazepine | 1.94% | 3.45% | 0% | — |
| Rifampin | 2.91% | 3.45% | 2.22% | — |
| Indication (VTE) | 64.71% | 82.75% | 40.91% | 47.04% |
Top 10 SNPs in discovery cohort analysis
| SNP . | Chromosome:position . | Gene . | P value (uncorrected) . |
|---|---|---|---|
| rs7856096 | chr9:130566539 | FPGS | 1.82 × 10−8 |
| rs11554716 | chr9:130565326 | FPGS | 2.93 × 10−7 |
| rs28694704 | chr17:18098253 | ALKBH5 | 6.56 × 10−6 |
| rs3750371 | chr11:50257362 | LOC441601 | 9.87 × 10−6 |
| rs2991344 | chr1:41978566 | HIVEP3 | 1.09 × 10−5 |
| rs2483689 | chr1:41978825 | HIVEP3 | 1.09 × 10−5 |
| rs82825 | chr3:52471942 | SEMA3G | 2.25 × 10−5 |
| rs7257009 | chr19:43866906 | CD177 | 2.47 × 10−5 |
| rs7406 | chr1:225589473 | LBR | 2.86 × 10−5 |
| rs1062098 | chr19:44930738 | ZNF229 | 2.87 × 10−5 |
| SNP . | Chromosome:position . | Gene . | P value (uncorrected) . |
|---|---|---|---|
| rs7856096 | chr9:130566539 | FPGS | 1.82 × 10−8 |
| rs11554716 | chr9:130565326 | FPGS | 2.93 × 10−7 |
| rs28694704 | chr17:18098253 | ALKBH5 | 6.56 × 10−6 |
| rs3750371 | chr11:50257362 | LOC441601 | 9.87 × 10−6 |
| rs2991344 | chr1:41978566 | HIVEP3 | 1.09 × 10−5 |
| rs2483689 | chr1:41978825 | HIVEP3 | 1.09 × 10−5 |
| rs82825 | chr3:52471942 | SEMA3G | 2.25 × 10−5 |
| rs7257009 | chr19:43866906 | CD177 | 2.47 × 10−5 |
| rs7406 | chr1:225589473 | LBR | 2.86 × 10−5 |
| rs1062098 | chr19:44930738 | ZNF229 | 2.87 × 10−5 |
The top 2 SNPs are in LD; no other FPGS SNPs were in LD with the top hit. Chromosome position according to Chr37.p13.
Manhattan plot of discovery cohort. Bonferroni cutoff of 3.22 × 10−7 for significance. The 2 top SNPs are in LD and are in FPGS.
Manhattan plot of discovery cohort. Bonferroni cutoff of 3.22 × 10−7 for significance. The 2 top SNPs are in LD and are in FPGS.
Distribution of associated risk allele across diverse populations. The risk allele, which is the ancestral allele, is most prevalent on the African continent. Map generated using Human Genome Diversity Project (HGDP) data through the HGDP Selection Browser.34,35
Gene-based analysis shows no novel associations
No single gene reached statistical significance after Bonferroni correction. The list of the top 10 genes can be found in supplemental Table 4.
No novel, previously undescribed nonsynonymous exonic variants were observed in VKORC1 in the discovery cohort. The only nonsynonymous exonic variant observed was rs7200749, with an allele frequency of 0.157. This variant had been previously reported as affecting warfarin resistance in a small cohort of South African descent; however, in our model, the variant had a P value of .311.36 For CYP2C9, we found no novel nonsynonymous variants. However, previously described coding variation was observed and was also genotyped, and a summary can be found in supplemental Table 5.
Replication cohort: rs7856096 replicates in an independent cohort of 372 African Americans on warfarin
We replicated our finding in a cohort of 372 African Americans with known stable dose but who were not selected for extreme dose phenotype (supplemental Figure 2). Key demographics of this group can be found in Table 1. Available clinical covariates included gender, age, height, weight, amiodarone use (0 or 1), and indication (VTE or not). We used a linear model of log2-transformed dose and available covariates from the discovery model to test whether rs7856096 was significant. Genome-wide SNP array data were only available for the University of Alabama at Birmingham samples, and in this subset of the data, the principal components did not significantly correlate with dose. For the genetic covariates used, the allele frequency of VKORC1 −1639G>A allele was 0.0941. For the CYP2C9 composite score, 14.5% of individuals had 1 variant and 2.1% had 2 variants. The tested SNP, rs7856096, had an allele frequency of 0.205. In this replication cohort of 372 individuals, the association of rs7856096 with warfarin dose remained significant (P = .0458). The coefficients for the covariates in this linear model can be found in supplemental Table 6. Because our replication model was missing 2 covariates from the discovery model (principal component 1 and aspirin use), we tested the reduced model on the discovery cohort (supplemental Table 7) and confirmed that rs7856096 remained significant in the discovery cohort using the reduced model.
rs7856096 predicts warfarin sensitivity in the IWPC dose equation
We tested the degree to which the new SNP, rs7856096, could improve the prediction of warfarin dose. In particular, we modeled the added value of this SNP to the IWPC pharmacogenetic dose equation in a combined cohort, consisting of both our discovery and replication cohorts totaling 476 individuals. The distribution of residual dose in this combined cohort is shown in supplemental Figure 4. Although the discovery cohort had been selected for extreme dose, the residual or unexplained dose was normally distributed. In the combined cohort, each minor allele of rs7856096 contributed to the −5.81 mg/week change in the predicted dose (P = 3.93 × 10−5). The percentage variance in dose explained by the IWPC dose equation alone was 30.0%, as measured by adjusted R2, and rs7856096 explained an additional 3.30 percentage points of the dose variability.
rs7856096 is a cis-acting eQTL for FPGS in LCLs
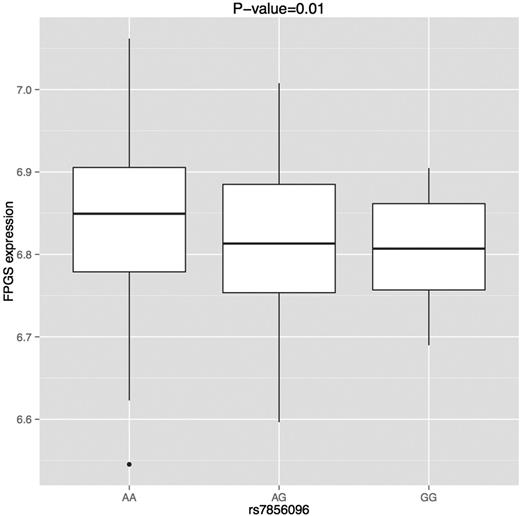
ENCODE data show that rs7856096 overlaps active enhancers in multiple cell types, including a lymphoblastoid cell line (GM12878) (supplemental Figure 5).28 This raises the possibility that allelic variants may affect enhancer activity and alter the expression of the target gene. We therefore evaluated the SNP for its association with the expression of FPGS in transformed lymphoblastoid cell lines derived from 3 HapMap African populations (MKK, LWK, and YRI). Transformed lymphoblastoid cell lines express FPGS to the same level as liver tissue (supplemental Figure 6).37 The SNP was significantly associated with the expression of the gene (P = .02; Figure 4). When we used the population indicator (which was significantly associated with FPGS expression at P = .007 in a univariate association analysis) as a covariate, the SNP remained significant (P = .01). The SNP had concordant direction of effect in all 3 African populations.
Effect of SNP on FPGS expression in LCLs across 3 African populations (MKK, LWK, and YRI). Effect on expression was evaluated using a linear model with population and rs7856096 (modeled additively) as covariates.
Effect of SNP on FPGS expression in LCLs across 3 African populations (MKK, LWK, and YRI). Effect on expression was evaluated using a linear model with population and rs7856096 (modeled additively) as covariates.
Discussion
Here, we describe a population-specific variant in a folate homeostasis gene that is associated with lower warfarin dose in African Americans. This variant not only adds to the list of variants influencing warfarin dose in African Americans but is also the first genetic evidence linking folate pathways to warfarin dosing.
Warfarin’s interindividual dose variability and narrow therapeutic index pose a challenge for accurate dosing. Dosing algorithms using currently available pharmacogenetic and clinical factors are 1 way to try and predict an individual’s dose.5 The COAG trial found no difference in percentage of time in the international normalized ratio when comparing patients dosed using a clinical algorithm (not currently standard of care) and patients dosed using a pharmcogenetic algorithm. However, the trial was underpowered to detect differences in adverse events, such as bleeding or thromboembolism.10 Additionally, another study found that pharmacogenetic algorithms can improve dose prediction compared with the current standard of care in the United Kingdom and Sweden.11 The COAG trial included 27% African Americans and concluded that using the genetic variants VKORC1 −1639G>A, CYP2C9*2, and CYP2C9*3 led to dose overprediction in this population compared with using clinical variables alone.9 However, these variants occur at a significantly lower frequency in African Americans, and other population-specific variants may improve prediction and improve performance in this population.38,39 In our study, we conditioned our model on the CYP2C9 variants that are more prevalent in African Americans, such as CYP2C9*5, *8, and *11, and discovered a novel variant affecting warfarin dose in this population.38 The resulting variant, rs7856096, is in a gene (and therefore pathway) not previously associated with the pharmacogenetics of warfarin.
Extreme phenotype design can help identify genetic factors affecting a phenotype when the discovery cohort is small.20,40,41 Because an extreme-phenotype approach coupled with sequencing has never been applied to the warfarin phenotype, our study was uniquely designed to find signals not detected in previous genome-wide association studies. After conditioning on known clinical factors and genetic factors (including CYP2C9 variants specific to African Americans), our approach yielded rs7856096, and this SNP was genome-wide significance after Bonferroni correction. This SNP is on chromosome 9, in the FPGS gene, which encodes a mitochondrial enzyme involved in folate homeostasis. rs7856096 is at the 5′ end upstream of exon 2 of FPGS. Within this data set, rs7856096 is in strong LD (D′ = 0.93) with another intronic SNP, rs11554716, but is not in LD with any other sequenced FPGS variants, leading to a sparse peak in the Manhattan plot. This variant was discovered because of its proximity to the coding region, and we found no coding variants in LD with it. The ancestral allele, which is the risk allele, is most prevalent in the African subcontinent, and would not likely have been observed in any of the studies in individuals of European descent.32 Additionally, the use of an extreme phenotype design allowed the identification of an association for which the signal may have been masked by the middle of the dose distribution. However, we did replicate this SNP in an independent cohort in which doses were not selected for an extreme phenotype.
In both the discovery and replication cohort analyses, the coefficient for the association between the minor rs7856096 allele and warfarin dose is negative, indicating that the presence of this variant is correlated with lower dose. We assessed the clinical value by calculating the information added to the IWPC dose equation using a combination of the discovery and replication cohorts. As mentioned in the methods, we included African American-specific CYP2C9 variants in this calculation, such as CYP2C9*5, *6, *8, and *11. We found in the combined cohort each risk allele corresponded to a dose reduction of 5.81 mg/week (0.83 mg/day). To truly assess the clinical impact of this variant and update the IWPC dose equation, additional studies in independent larger cohorts are required. However, this first clinical analysis suggests that individuals homozygous for the risk allele could require a clinically significant reduction of 11.62 mg/week (1.66 mg/day) from the IWPC predicted dose. In our combined cohort, 3.3% of individuals were homozygous at this locus. For comparison, currently, the Food and Drug Administration label on warfarin recommends a reduction of 2 to 4 mg/day for wild-type VKORC1 with CYP2C9 *1/*3, *2/*2, or *2/*3 and no reduction for CYP2C9 *1/*2.42
Preliminary evidence from 3 HapMap African populations suggests that the action of this SNP is through the regulation of FPGS expression. Changes in FPGS expression are known to affect folate sequestration and utilization in the folate pathway.43,44 This analysis was based on transformed lymphoblastoid cell lines because African liver samples were not available; however, RNA-Seq data in European populations show that transformed lymphoblastoid cell lines express FPGS at the same level as liver tissue (supplemental Figure 6).37 FPGS is expressed and active in the liver, the primary site of warfarin metabolism (supplemental Figure 7).45,46 From a biological standpoint, the discovery of this variant is the first genetic evidence of a connection between the warfarin and folate pathways. Previously, folate had been linked to warfarin through drug-drug interaction studies—folate intake has been shown to affect warfarin clearance, and a small study demonstrated that increased folate intake affected warfarin metabolism rates.47 Additionally, folate has been shown to affect coagulation status, so this variant could affect the warfarin pharmacodynamic pathway as well.48,49 In this study, we had no information on folate intake; however, one could postulate that, because this variant affects folate sequestration, a possible genetic-environmental interaction could occur. Studies looking at how this variant and folate interact in the setting of warfarin could shed light on any possible synergistic effects.
In conclusion, we present the first report of a SNP involved in folate homeostasis, which significantly influences warfarin dose in African Americans, a population whose dose variability has been understudied.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
Work by L.H.C. was supported by American Heart Association Midwest Affiliate Spring 2010 grant-in-aid 10GRNT3750024 and a University of Illinois at Chicago College of Pharmacy Hans Vahlteich research award. Work by N.L. was supported in part by National Heart Lung and Blood Institute grant RO1HL092173 and National Centre for Advancing Translational Sciences grant UL1 TR000165. Work by J.A.J. and T.L. was supported in part by National Institutes of Health, National Institute of General Medical Sciences grant U01 GM074492. Work by M.A.P. was supported in part by National Heart Lung and Blood Institute grant K23 HL089808 and American Heart Association grant 10CRP3740026. Work by R.B.A. and T.E.K. is supported in part by National Institutes of Health, National Institute of General Medical Sciences grant GM61374 and gifts from Microsoft and Lightspeed Ventures. Work by R.D. was supported in part by the Howard Hughes Medical Institute Medical Fellows Program, the Stanford Medical Scientist Training Program, and the Stanford Genetics Training Program. Illumina sequencing services were performed by the Stanford Center for Genomics and Personalized Medicine.
Authorship
Contribution: R.D., E.R.G., L.H.C., J.A.J., T.E.K., S.H., B.P., K.J.K., C.D.B., R.B.A., and M.A.P. helped design the research; E.R.G., B.B., L.H.C., J.A.J., N.L., T.L., S.R.P., and M.A.P. helped generate the data; R.D., E.R.G., S.H., B.P., K.J.K. helped analyze the data; R.D. and E.R.G. drafted the manuscript; and all authors edited the manuscript.
Conflict-of-interest disclosure: L.H.C. is a coinvestigator for US Utility Patent application no. 12/572 908, titled “CYP2C9*8 alleles correlate with decreased warfarin metabolism and increased warfarin sensitivity”: published May 27, 2010; Pub. No. US 2010/0130599. R.B.A. is a founder of and consultant for Personalis. C.D.B. is on the advisory board of a project at 23andMe and on the scientific advisory boards of Personalis, Inc.; InVitae; Etalon, Inc.; and Ancestry.com. T.E.K. is a consultant for Personalis. The remaining authors declare no competing financial interests.
The current affiliation for L.H.C. is Department of Pharmacotherapy and Translational Research, University of Florida, Gainesville, FL.
Correspondence: Russ Altman, 443 Via Ortega, Room 209, MC: 4245, Stanford, CA 94305-4125; e-mail: russ.altman@stanford.edu; or Minoli Perera, 900 E 57th St, Chicago, IL 60637; e-mail: mperera@bsd.uchicago.edu.
References
Author notes
R.D. and E.R.G. contributed equally to this work.
R.B.A. and M.A.P. contributed equally to this work.