Abstract
Intra-tumoral genetic heterogeneity and clonal evolution play a central role in disease relapse, and constitute one of the foremost obstacles to cancer cure. To measure intra-tumoral genetic heterogeneity, we recently developed an analytic framework which utilizes whole-exome sequencing (WES) to calculate the proportion of cancer cells within a sample harboring each mutation. Applying this approach to a clinically heterogeneous cohort of chronic lymphocytic leukemia (CLL), we previously discovered that the intra-tumoral genetic heterogeneity in primary patient samples prior to treatment initiation anticipates clonal evolution and is linked to adverse clinical outcome with therapy (Landau et al., Cell 2013).
To definitively evaluate the value of intra-tumoral heterogeneity in predicting clinical outcome in CLL, we performed an analogous analysis of pre-treatment samples collected from study subjects enrolled on the CLL8 trial of the GCLLSG (Hallek et al., Lancet 2010). In this multicenter phase III study, 817 patients were randomized to receive as first-line therapy fludarabine and cyclophosphamide either alone or in combination with rituximab. Paired pre-treatment tumor DNA from CD19+ selected peripheral blood mononuclear cells (PBMC), and non-tumor DNA from CD19- PBMC were available from 309 subjects (61% IGHV unmutated, 26% del(11q), 4.5% del(17p)). From these 309 sample pairs, we performed SNP array analysis and WES. Somatic mutations were identified as single nucleotide variants (SNVs), indels and copy number alterations (CNAs), detected in the tumor DNA but not in matched germline DNA. Subsequently, we utilized the algorithm ABSOLUTE to calculate the purity and ploidy of each sample, and classify every mutation as either clonal (i.e., present in all leukemic cells in the sample) vs. subclonal (i.e., present in a subset of leukemic cells). Progression free survival (PFS) was defined as the primary outcome measure.
Preliminary sequencing results for 275 of 309 CLL samples are currently available. Across these samples, WES identified 7290 somatic SNVs and indels. Overall mutation rate for these samples was 0.8/Mb, similar to prior large-scale reports of DNA sequencing of CLL. By applying the algorithm MutSig, which identifies putative driver genes recurrently mutated more than expected by chance, we detected 16 of 24 previously described CLL drivers (e.g., TP53, ATM, SF3B1 and NOTCH1) to significantly affect this cohort. In addition, due to the large sample size we have identified 9 novel putative drivers in CLL including MGA, IKZF3 and ARID1A. CNA analysis identified the previously reported recurrent CNAs including (del(13q), del(11q), del(17p), trisomy 12 and del(8p)).
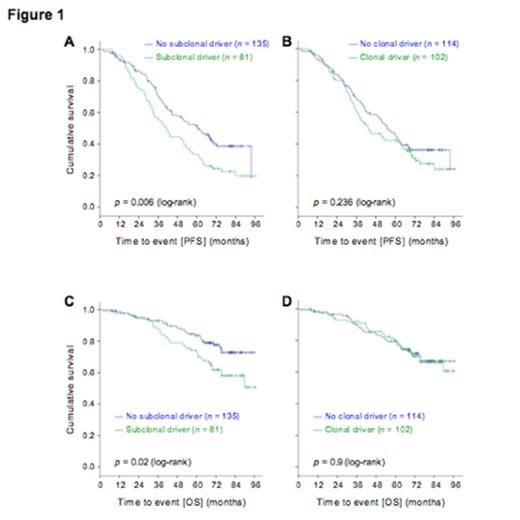
At this time, subclonal analysis by ABSOLUTE is available for 216 of 309 CLLs. Across the 216 samples, we identified 2041 clonal and 2106 subclonal mutations. A putative driver gene mutation was identified as clonal in 102 (47.2%) patients, and as subclonal in 81 (37.5%) patients. The presence of a clonal driver was not significantly associated with differences inPFS (logrank P= 0.236). In contrast, the presence of a subclonal driver was associated with a significantly shortened PFS (logrank P= 0.006, Figure 1A-B). A Cox regression model that included the presence of a subclonal driver as well as the treatment arm assignment (FC vs. FCR), retained the presence of a subclonal driver as statistically significant in association with shorter PFS (HR 1.56 [95%CI 1.12-2.19], P=0.009). The presence of a subclonal driver was furthermore associated with shorter overall survival (logrank P=0.02, Fig 1C-D).
This preliminary analysis demonstrates that pre-treatment characterization of intra-tumoral genetic heterogeneity can identify patients at risk for inferior outcome with first-line fludarabine-based therapy in CLL. With completion of this analysis for the entire cohort of 309 patients, we plan to perform a multivariable analysis to evaluate the clinical impact of clonal and subclonal driver mutations in CLL. Further ongoing studies include a longitudinal genetic evaluation of the subset of patients with subsequent disease relapse in order to unravel the evolutionary dynamics underlying disease progression.
Fischer:Roche: Travel grants Other. Hallek:Janssen, Pharmacyclics: Consultancy, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal