Abstract
Gene expression profiling (GEP) via microarray analysis is a widely used tool for assessing risk and other patient diagnostics in clinical settings. However, non-biological factors such as systematic changes in sample preparation, differences in scanners, and other potential batch effects are often unavoidable in long-term studies. In order to reduce the impact of batch effects on microarray data, Johnson et al developed ComBat for use when combining batches of gene expression microarray data. However, ComBat adjusts samples to the overall mean--this may prevent GEP-models designed on historical samples to function properly on ComBat-adjusted data. We propose a modification that centers all samples to the location and scale of an unadjusted, gold standard' batch. This modified ComBat (M-Combat) simplifies the inclusion of external validation sets or, in our case, the rescaling of baseline Myeloma GEP samples to a desired, historical standard to allow for previous research (GEP70, GEP80, molecular subgroup, etc.) to continue to function properly. We illustrate M-ComBat by transforming baseline purified plasma cell GEP samples from patients with Multiple Myeloma (MM) enrolled in UARK Total Therapy 2, 3, 4, 5, and 6 across three main batches.
Baseline purified plasma cells were gathered from patients enrolled on different total therapies and processed on Affymetrix U133Plus 2.0 microarrays between 2004 and 2014. These samples were analyzed by scanners at two different laboratorieseither at the Myeloma Institute or, beginning in 2011, at Signal Genetics. Samples were prepared with either the One-Cycle and Two-Cycle Target Labeling and Control Reagents (old' kit) or, beginning in 2009, the 3' IVT Express Kit (new' kit). We standardize baseline GEP data from three main batches (old' kit Myeloma Institute, new' kit Myeloma Institute, and new' kit Signal Genetics) to the old' kit Myeloma Institute standard so that research from the historical standard such as the GEP70, GEP80, and molecular subgroup calculator continue to function after performing M-ComBat.
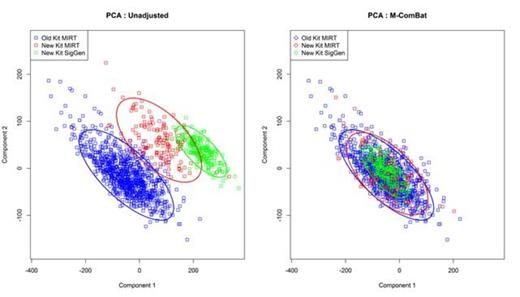
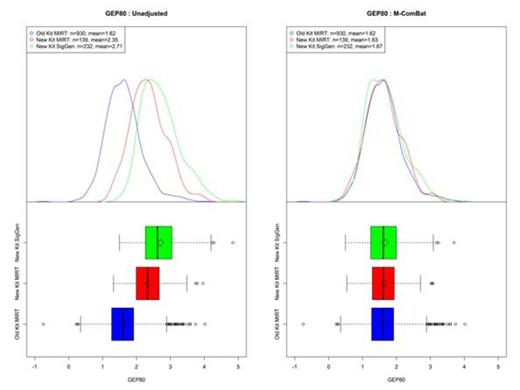
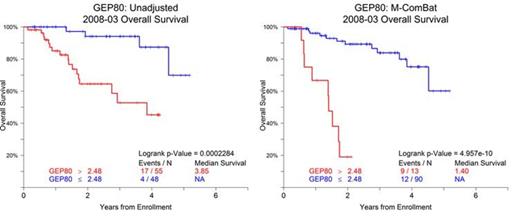
M-ComBat reduced the batch-effect related significant differences while shifting samples to the desired standard. When visualizing the top two principal components of the full GEPs of the baseline MM samples, the M-ComBat transformed data is shifted to the old' kit Myeloma Institute centroid (Figure 1). The GEP80 risk score is susceptible to batch effects; however after using M-ComBat, the distribution of GEP80 scores increase in agreement across batches (Figure 2). The separation in outcomes of GEP80 high and low risk groups is more evident after performing M-ComBat (unadjusted p-value: 2.28e-04 & M-ComBat p-value: 4.96e-10, Figure 3). Other GEP-based metrics such as the molecular subgroup calculator also increased in agreement. At the 0.1 significance level, the unadjusted data showed dependency between sample preparation and molecular subgroup (Chi-square test p-value : 0.071), while the M-ComBat adjusted data showed independence (Chi-square test p-value : 0.477). Thus after using M-ComBat, the distribution of subgroups (CD1, CD2, HY, LB, MF, MS, or PR) better reflected the distribution of historical samples. Other investigations illustrated the ability of M-ComBat to improve agreement of other GEP-based scores and revealed the greater power to see true biological differences.
M-ComBat is a practical modification to an accepted method that adjusts diverse data to a pre-determined standard of samples with ease. This method is valuable for a variety of situations including ours, in which GEP samples were analyzed over a ten year span and batch effects were unavoidably introduced. M-ComBat allows for previous risk models to remain viable on future batches of samples--thus eliminating the need for redefining cut-points or modifying models.
Principal Components Analysis. Scatter plot of the top two components from Principal Components Analysis (PCA) of the pre and post M-ComBat transformed baseline purified plasma cell sample GEPs.
Principal Components Analysis. Scatter plot of the top two components from Principal Components Analysis (PCA) of the pre and post M-ComBat transformed baseline purified plasma cell sample GEPs.
GEP80 for Baseline MM Purified Plasma Cell Samples. Boxplot and density curves of GEP80 scores for all baseline MM purified plasma cell samples both unadjusted and M-ComBat shifted.
GEP80 for Baseline MM Purified Plasma Cell Samples. Boxplot and density curves of GEP80 scores for all baseline MM purified plasma cell samples both unadjusted and M-ComBat shifted.
Overall Survival by GEP80 for Baseline MM Purified Plasma Cell Samples from Total Therapy 6 (UARK 2008-03). Kaplan-Meier Survival curve for High and Low risk defined by GEP80.
Overall Survival by GEP80 for Baseline MM Purified Plasma Cell Samples from Total Therapy 6 (UARK 2008-03). Kaplan-Meier Survival curve for High and Low risk defined by GEP80.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal