Abstract
In many parts of the world, diagnosis and monitoring of CML patients is limited by the availability and cost of molecular testing. In countries without molecular diagnostic capabilities, blood samples can be shipped to central labs, but this is both hampered by sample degradation, and the high costs of shipping. This study explores the method of directly spotting peripheral blood onto a paper template (dried blood spots), with subsequent shipping, RNA extraction, and BCR-ABL testing.
Methods:
Blood Spots and Shipment. We received dried blood spots from Australia and African countries by mail or courier, and blood from CML patients from our institution were also used for these experiments. 200μL of blood (PB) was pipetted onto Whatman 503 Protein Saver Cards (PSC; Sigma-Aldrich), where each card contains four 50μL spots. Cards were allowed to dry for at least 24 hours at room temperature. For mailing, PSCs were sealed into glassine envelopes with a packet of desiccant, and then placed inside a mailing envelope following DOT and IATA regulation for shipping non-regulated, exempt human specimens.
RNA Extraction from Cards and %BCR-ABL determination. Blood spots were incubated with proteinase K followed by RNA isolation using RNeasy Mini Kits (Qiagen). Extracted RNA was quantified using a NanoDrop spectrometer (Thermo Scientific). %BCR-ABL was determined using the automated Cepheid GeneXpert platform or manual two-step quantitative RT-PCR on the 7900HT Fast Real-Time PCR System (Applied Biosystems).
Results:
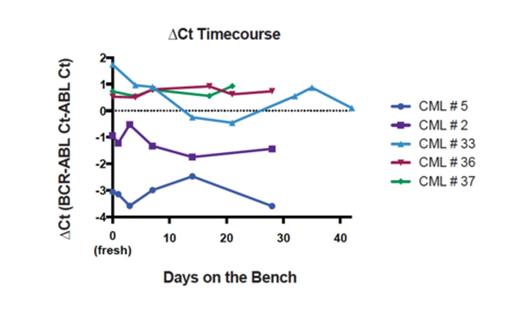
Bench top time course: To test for effects of long transit times on RNA quality, we performed a time course study of cards at room temperature (RT) with 5 samples. For each sample, multiple cards were spotted with PB. The cards were then allowed to sit at RT for predetermined amounts of time, up to 42 days, before extracting RNA. We measured RNA integrity for one of the specimens (CML # 5) and found rapid degradation with the RIN number going from 8.7 for the fresh blood to 2.8 after 28 days on the card. However the amplification for both BCR-ABL and ABL differed less than one cycle between the fresh blood and the last time point by manual qRT-PCR (BCR-ABL Ct = 23.63 for fresh blood and 24.06 for day 28 PSC; ABL Ct = 26.69 for fresh blood and 27.64 for day 28 PSC). Figure 1 shows the results of the time course experiment for the 5 samples as a plot of ΔCt versus time in days.
BCR-ABL qRT-PCR concordance studies: We compared the %BCR-ABL results obtained in fresh specimen at the institution sending the sample with the %BCR-ABL results we obtained from RNA extracted from PSC using the Cepheid GeneXpert. Paired evaluable results were available for 9 samples with a median WBC = 9.8 x 109/L (range: 3.37x109/L – 85.5x109/L). Samples were 8 to 49 days old at the time of extraction. The amount of RNA input into the GeneXpert reaction ranged from 38.75ng to 1μg. The %BCR-ABL detected ranged from 0.37% to 27% (see Table). The mean absolute difference between fresh blood and PSC BCR-ABL% is 2%; the relative mean percent change for BCR-ABL, using fresh blood as the reference is 13.1% (S.D., 31.2), P = 0.24.
Conclusions and future directions:
Dried blood spots are relatively inexpensive method to transport blood that preserves enough RNA stability to allow highly accurate BCR-ABL detection, when compared to results performed on an identical platform using fresh peripheral blood samples.
Further studies are undergoing to accurately determine the sensitivity of this method and the feasibility of using regular mail for inexpensive transport of specimens.
| ID . | WBC (1000/μL) . | Sample Age at Spotting (Days) . | Sample Age at RNA extraction (Days) . | RNA ng/μl . | Volume GeneXPert (μL) . | Paper %BCR-ABL (IS) GeneXpert . | Fresh Blood % BCR-ABL (IS) GeneXpert . |
|---|---|---|---|---|---|---|---|
| I1 | na | 0 | 10 | 426 | 3 | 49 | na |
| I2 | 24.1 | 0 | 13 | 110 | 9 | 27 | 45 |
| I3 | 80 | 0 | 9 | 18 | 15 | 44 | na |
| I4 | 7.4 | 2 | 8 | 5 | 10 | 2.4* | 3.1 |
| I5 | 5.5 | 0 | 49 | 5 | 24 | 1.9 | 2 |
| I6 | 3.6 | 1 | 30 | 7.42 | 25 | 9 | 12 |
| I7 | 85.5 | 1 | 30 | 102 | 10 | 24 | 39 |
| I8 | 12.2 | 1 | 29 | 12.4 | 15 | 12 | 8.8 |
| I9 | na | 1 | 28 | 1.5 | 25 | 0.37* | 0.71 |
| I10 | 3.37 | 0 | 27 | 3 | 25 | 7.8 | 5.7 |
| I11 | 15.9 | 1 | 27 | 31 | 10 | 23 | 25 |
| I12 | 6.6 | 1 | 27 | 14.4 | 15 | na | 2.3 |
| ID . | WBC (1000/μL) . | Sample Age at Spotting (Days) . | Sample Age at RNA extraction (Days) . | RNA ng/μl . | Volume GeneXPert (μL) . | Paper %BCR-ABL (IS) GeneXpert . | Fresh Blood % BCR-ABL (IS) GeneXpert . |
|---|---|---|---|---|---|---|---|
| I1 | na | 0 | 10 | 426 | 3 | 49 | na |
| I2 | 24.1 | 0 | 13 | 110 | 9 | 27 | 45 |
| I3 | 80 | 0 | 9 | 18 | 15 | 44 | na |
| I4 | 7.4 | 2 | 8 | 5 | 10 | 2.4* | 3.1 |
| I5 | 5.5 | 0 | 49 | 5 | 24 | 1.9 | 2 |
| I6 | 3.6 | 1 | 30 | 7.42 | 25 | 9 | 12 |
| I7 | 85.5 | 1 | 30 | 102 | 10 | 24 | 39 |
| I8 | 12.2 | 1 | 29 | 12.4 | 15 | 12 | 8.8 |
| I9 | na | 1 | 28 | 1.5 | 25 | 0.37* | 0.71 |
| I10 | 3.37 | 0 | 27 | 3 | 25 | 7.8 | 5.7 |
| I11 | 15.9 | 1 | 27 | 31 | 10 | 23 | 25 |
| I12 | 6.6 | 1 | 27 | 14.4 | 15 | na | 2.3 |
*%BCR-ABL was manually calculated due to late ABL Cts because of low starting material.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.
This icon denotes a clinically relevant abstract

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal