Abstract
Background: Ex-vivo expansion of CBT-cells using CD3/CD28 co-stimulatory beads, IL-2 + IL-7 and subsequent priming against leukemia cell lines using IL-15 generated specific CTLs. [1, 2]
Hypothesis: We hypothesized that (a) patient-derived AML-specific PB auto CTLs could be generated with immune-stimulatory culture condition (b) Resistant AML samples would possess gene expression profiles similar to MDSCs (myeloid-derived suppressor cells) (c) Frequency of Tregs (CD4+CD25brightFoxP3+) and T-cell co-signaling molecules gene expression will be different between effective and ineffective CTLs.
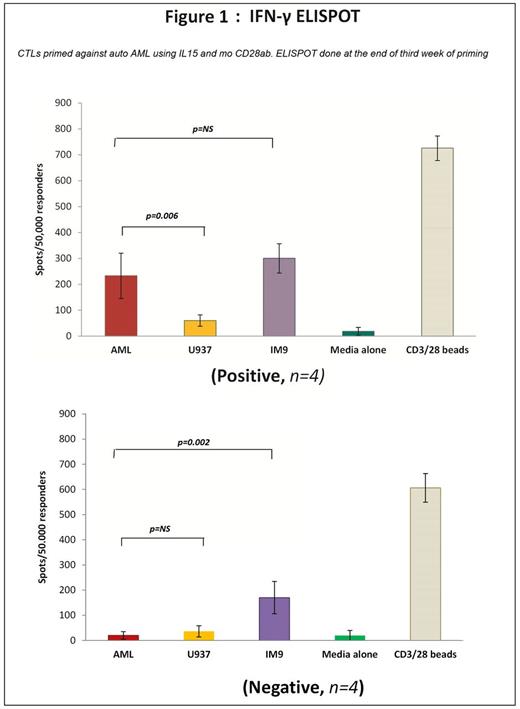
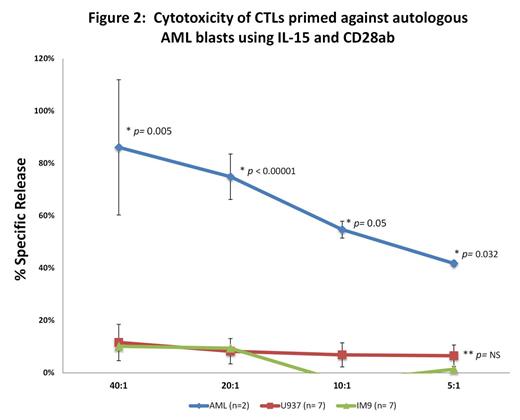
Methods: AML & auto T-cells were purified from cryopreserved PBMC of AML patients admitted with acute blast crisis (n=8). AML blasts were sustained in StemSpan™ Serum-Free media [STEMCELL Technologies] with MSC support + cytokine cocktail (IL-3, SCF, FLT3L, GMCSF, IL-4). T-cells were expanded in culture for 2 weeks as reported [1, 2] and subsequently primed with γ-irradiated auto AML weekly X 3 with IL15 + CD28ab [BD Biosciences]. At the end of week 3 (EOW3), cytotoxicity was assessed against AML and irrelevant targets - IM9 (lymphoid) and U937 (myeloid) cell lines, loaded with BATDA at an E:T ratio of 40:1, 20:1, 10:1 and 5:1 using DELFIA® EuTDA assay.[2] IFN-γ ELISPOT assay against same targets was also done.[2] RT-qPCR analysis was performed on AML & T-cells before and after priming, using Power SYBR® Green master mix (Thermo Fisher Scientific) and StepOne Plus system [Life Technologies]. Two-tailed student t-testcompared experimental groups.
Results
· T-cells expanded in all samples (n=8) with a median expansion of 155-fold (range 11-489), at EOW3.
· ELISPOT assay was positive in 4/8 samples. [Fig 1]
· CTL assay was difficult to standardize for primary AML blasts due to high degree of spontaneous apoptosis (>30% spontaneous release [SR]).
· 2/8 samples were deemed evaluable (SR<30%).
· Both samples showed AML-specific lysis. [Fig 2]
· Overall, AML-specific autologous CTL could be generated from 5 of 8 samples based on ELISPOT & CTL assays, regardless of original FAB immunophenotype, not shown.
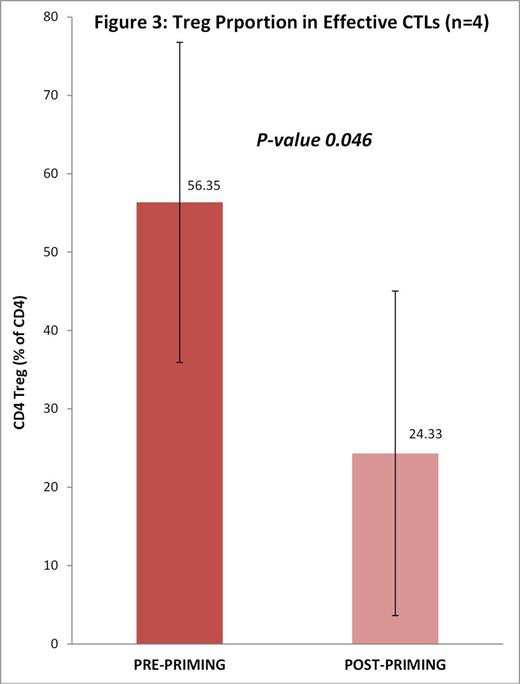
· Tregs proportion declined significantly in effective CTLs post-priming as compared to pre-priming (56% to 24%, p-value 0.046, n=4). [Fig 3]
· T-cell gene expression profiling showed significant differences in effective vs ineffective CTLs. [Table 1]
· Resistant AML (n=3) had up-regulated downstream markers associated with MDSC generation compared to “non-resistant” AML (n=5). [Table 2]
Conclusions (a) AML-specific auto CTLs can be generated (b) Tregs decreased with priming in effective CTLs (c) differential T-cell gene expression profile exists between effective and ineffective CTLs (d) AML gene expression suggests MDSC-like profile in resistant samples.
T-CELL GENE EXPRESSION PROFILE (POST VS PRE-PRIMING)
| Effective CTLs (n=5) | Ineffective CTLs (n=3) | |||||
| Gene | ΔΔ Ct (Post - Pre) (mean, SEM) | P-value | Fold change (mean, SEM) | ΔΔ Ct (Post - Pre) (mean, SEM) | P-value | Fold change (mean, SEM) |
| 4-1BB | -3.17 (0.76) | 0.025 | 14 (7.7) | 1.98 (1.04) | 0.19 | 0.39 (0.22) |
| HVEM | -2.43 (0.61) | 0.028 | 7.3 (3.7) | 0.14 (1.65) | 0.95 | 1.57 (1.28) |
| LIGHT | -3.62 (0.73) | 0.016 | 17.3 (7.3) | 1.78 (1.84) | 0.44 | 1.1 (0.98) |
| PRKC-α | -2.03 (0.47) | 0.023 | 4.6 (1.1) | 1.89 (0.36) | 0.034 | 0.29 (0.08) |
| PRKC-θ | -3.36 (0.59) | 0.01 | 13.7 (6.7) | 0.25 (0.59) | 0.71 | 0.99 (0.41) |
| LAIR1 | -3.81 (0.42) | 0.003 | 16.2 (5.6) | -1.35 (2.20) | 0.60 | 17.15 (16.5) |
| PP2A | -2.40 (0.57) | 0.025 | 6.7 (2.6) | 0.49 (1.57) | 0.79 | 1.89 (1.52) |
| 2B4 | -1.53 (1.14) | 0.27 | 4.98 (1.82) | -3.48 (0.11) | 0.02 | 11.2 (0.9) |
| LTA-α | -1.18 (0.78) | 0.23 | 3.61 (2.11) | 2.69 (0.18) | 0.043 | 0.16 (0.02) |
| LTA-β | -0.93 (0.63) | 0.24 | 2.49 (0.99) | 2.24 (0.47) | 0.042 | 0.23 (0.08) |
| Effective CTLs (n=5) | Ineffective CTLs (n=3) | |||||
| Gene | ΔΔ Ct (Post - Pre) (mean, SEM) | P-value | Fold change (mean, SEM) | ΔΔ Ct (Post - Pre) (mean, SEM) | P-value | Fold change (mean, SEM) |
| 4-1BB | -3.17 (0.76) | 0.025 | 14 (7.7) | 1.98 (1.04) | 0.19 | 0.39 (0.22) |
| HVEM | -2.43 (0.61) | 0.028 | 7.3 (3.7) | 0.14 (1.65) | 0.95 | 1.57 (1.28) |
| LIGHT | -3.62 (0.73) | 0.016 | 17.3 (7.3) | 1.78 (1.84) | 0.44 | 1.1 (0.98) |
| PRKC-α | -2.03 (0.47) | 0.023 | 4.6 (1.1) | 1.89 (0.36) | 0.034 | 0.29 (0.08) |
| PRKC-θ | -3.36 (0.59) | 0.01 | 13.7 (6.7) | 0.25 (0.59) | 0.71 | 0.99 (0.41) |
| LAIR1 | -3.81 (0.42) | 0.003 | 16.2 (5.6) | -1.35 (2.20) | 0.60 | 17.15 (16.5) |
| PP2A | -2.40 (0.57) | 0.025 | 6.7 (2.6) | 0.49 (1.57) | 0.79 | 1.89 (1.52) |
| 2B4 | -1.53 (1.14) | 0.27 | 4.98 (1.82) | -3.48 (0.11) | 0.02 | 11.2 (0.9) |
| LTA-α | -1.18 (0.78) | 0.23 | 3.61 (2.11) | 2.69 (0.18) | 0.043 | 0.16 (0.02) |
| LTA-β | -0.93 (0.63) | 0.24 | 2.49 (0.99) | 2.24 (0.47) | 0.042 | 0.23 (0.08) |
GENE EXPRESSION PROFILE RESISTANT VS NON-RESISTANT AML
| Gene | ΔΔ Ct (mean, SEM) | 95% CI | P-value | Relative fold change | |
| JAK1 | -4.63 (1.98) | -9.48 | 0.21 | 0.0579 | 24.83 |
| JAK2 | -5.38 (0.94) | -7.67 | -3.08 | 0.0012 | 41.52 |
| JAK3 | -5.90 (2.17) | -12.81 | 1.01 | 0.0726 | 59.77 |
| S100A8 | -7.16 (2.66) | -14.01 | -0.32 | 0.0432 | 143.27 |
| S100A9 | -8.31 (2.75) | -15.04 | -1.59 | 0.0233 | 318.37 |
| c-myc | -2.78 (0.59) | -4.24 | -1.33 | 0.0034 | 6.89 |
| Gene | ΔΔ Ct (mean, SEM) | 95% CI | P-value | Relative fold change | |
| JAK1 | -4.63 (1.98) | -9.48 | 0.21 | 0.0579 | 24.83 |
| JAK2 | -5.38 (0.94) | -7.67 | -3.08 | 0.0012 | 41.52 |
| JAK3 | -5.90 (2.17) | -12.81 | 1.01 | 0.0726 | 59.77 |
| S100A8 | -7.16 (2.66) | -14.01 | -0.32 | 0.0432 | 143.27 |
| S100A9 | -8.31 (2.75) | -15.04 | -1.59 | 0.0233 | 318.37 |
| c-myc | -2.78 (0.59) | -4.24 | -1.33 | 0.0034 | 6.89 |
Refs:
1.Davis et al. Cancer Research 2010;70(13):5249
2.Jeyaraj A, Chen X, Szabolcs P. IL-15 Induced Polyclonal CTL Generated From Expanded CBT Cells Against Leukemia Cell Lines Constitutes IFN-γ Producing Cells and TCRγδ Cells. ASH 2012 Annual Meeting
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal