Key Points
STAT3 directly regulates expression of NKG2D in NK cells.
Defects in STAT3 signaling result in deficient NKG2D responses to cytokine.
Abstract
Signal transducer and activator of transcription 3 (STAT3) is considered a negative regulator of inflammation, as inhibition of STAT3 signaling enhances antitumor immunity. However, STAT3 activation is a key oncogenic pathway in natural killer (NK)-lineage large granular lymphomas, and we recently reported enhanced proliferation and function of human NK cells activated with IL-21, which signals primarily through STAT3. These IL-21–expanded NK cells also have increased NKG2D expression, which led us to focus our investigation on whether STAT3 regulates NKG2D. In this study, we show that modulation of STAT3 phosphorylation with cytokines and small-molecule inhibitors correlates with NKG2D expression on human NK cells, leading to altered NK-cell degranulation. Moreover, NKG2D expression on murine NK cells having conditional STAT3 ablation is lower than on NK cells from wild-type mice, and human NK cells carrying dominant-negative STAT3 mutations have decreased baseline NKG2D expression and blunted responses to IL-10 and IL-21. Lastly, we show binding of STAT3 to a predicted STAT3 binding site upstream of the NKG2D gene, which is enhanced by IL-10 and IL-21 and decreased by STAT3 inhibition. Taken together, these data show that NKG2D expression in NK cells is regulated at the transcriptional level by STAT3, resulting in a functional NK cell defect in patients with STAT3 mutations.
Introduction
Signal transducer and activator of transcription 3 (STAT3) is a pleiotropic transcription factor that transmits signals from the extracellular environment to the nucleus, mediating downstream signaling of many cytokines. STAT3 is recruited to the activated cytokine receptor and then tyrosine-phosphorylated by receptor-associated janus kinase (JAK). Upon phosphorylation, STAT3 molecules dimerize by reciprocal interaction of their phosphorylated SH2 domains, which in turn promotes translocation to the nucleus and binding to specific DNA elements to regulate transcription of target genes involved in proliferation, apoptosis, and differentiation.1 Constitutive STAT3 phosphorylation is common in cancer, and thus, STAT3 inhibitors are increasingly being tested in preclinical studies and clinical trials.2
STAT3 is also an important modulator of innate and adaptive immune responses. STAT3 mediates signal transduction for several cytokine families including common γ-chain cytokines (IL-2, IL-7, IL-15, and IL-21), the IL6/gp130 family (IL-6 and IL-27), interferons, IL-10, IL-12, and IL-23, and colony stimulating factors. STAT3 signaling is required for the generation and maintenance of Th17 cells,3 functional maturation of memory T cells,4 and T-cell–dependent differentiation of B cells into plasma cells.5 Dominant negative STAT3 mutants cause an immunologic deficiency (Job’s or Hyper-IgE Syndrome [HIES]), characterized by recurrent bacterial skin and lung infections.6 We previously demonstrated a proinflammatory role for STAT3 activation in maintaining neutrophil number and function.7,8 By contrast, inhibition of STAT3 in murine models improves antitumor immunity.9
Natural killer (NK) cells play a crucial role in immune response to viruses and tumors, destroying virally infected cells and neoplasms. Activating receptors, which recognize ligands that are increased on “stressed” target cells, transmit signals to activate cytolytic activity of NK cells. NKG2D is an activating receptor on NK cells that recognizes ligands induced by cellular stress such as heat shock, DNA damage, transformation, and viral and bacterial infection. Not surprisingly, NKG2D plays a critical role in the immune response mediated by NK cells to infections and tumors.10 Although much is known about the regulation of NKG2D ligands,11 little is known about the mechanisms of NKG2D receptor regulation.
NKG2D expression on NK cells is upregulated in response to IL-2, IL-15, IL-12, and INF-α,12,13 all of which predominantly signal through various STAT family members. Previous work from our laboratory showed that IL-21, which signals primarily through STAT3 in NK cells,14 is important in regulating survival and proliferation of NK cells through telomere maintenance.15 As NKG2D is a key receptor involved in NK-cell–mediated antitumor responses, we hypothesized that STAT3 activation may regulate NKG2D expression and NK-cell antitumor activity.
Materials and methods
Cells and cell lines
Anonymized normal donor (ND) buffy coats were obtained from the Gulf Coast Regional Blood Center (Houston, TX) under a protocol approved by the Institutional Review Board (IRB) of University of Texas MD Anderson Cancer Center. Peripheral blood was obtained from HIES patients at the National Institute of Allergy and Infectious Diseases and Children’s Hospital of Philadelphia under protocols approved by the IRB of each respective institution. IRB approval was obtained by J.S.O. and A.F.F./S.M.H. to acquire patient blood samples for immunologic research. IRB approval was obtained by D.A.L. to acquire samples from collaborators and perform this research. This study was conducted in accordance with the Declaration of Helsinki.
Peripheral blood mononuclear cells (PBMCs) were purified by centrifugation over Ficoll-Paque from healthy donor buffy coat samples and HIES patient blood samples. Fresh NK cells were purified from PBMCs by enriching to ≥ 95% purity (CD3−CD16/56+) with RosetteSep Human NK Cell Enrichment Cocktail (STEMCELL Technologies, Vancouver, BC, Canada).16
K562-based artificial antigen presenting cells (aAPCs) were produced by genetic modification of parental K562 to express CD64, CD86, CD137L, truncated CD19, and both membrane-bound IL-21 or IL-15.15 NK cells were expanded from PBMCs in vitro by weekly stimulation with the indicated aAPCs in the presence of 50 IU/mL of rhIL-2 as described previously.15 NK cells were purified as described above to a CD3+ content <1%16 prior to gene expression analysis.
Murine splenocytes were obtained from wild-type (WT) mice or mice in which floxed STAT3 was deleted in hematopoietic cells by Cre expression under the Tie2 promoter.8 Murine NK cells were isolated from splenocytes using EasySep Mouse NK Cell Enrichment Kit (STEMCELL Technologies). Institutional Animal Care and Use Committee approval was obtained by S.S.W. to conduct immunologic research in STAT3-deficient mice.
Cells were cultured in RPMI 1640 (Cellgro/Mediatech, Manassas, VA) supplemented with 10% fetal bovine serum (HyClone, Logan, UT), 2 mM l-glutamine (Gibco/Invitrogen, Carlsbad, CA), and 1% penicillin/streptomycin (Cellgro/Mediatech) with or without cytokines, as indicated.
Antibodies.
Murine monoclonal antibodies against human CD3, CD56, CD107a, NKG2D, antibodies against murine CD3, NKG2D, and NK1.1, and isotype control mAb were obtained from BD Biosciences (San Jose, CA). Antibodies against pSTAT1 (Tyr 701), total STAT3, pSTAT3 (Tyr 705), pSTAT5 (Tyr 694), and β-actin were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA).
Reagents.
JSI-124 and S3I-201 were purchased from Calbiochem (Gibbstown, NJ), and were dissolved in dimethylsulfoxide. Recombinant human IL-2 (Proleukin) was purchased from Novartis Vaccines and Diagnostics (East Hanover, NJ), IL-10 and IL-21 from PeproTech (Rocky Hill, NJ), and 7-AAD from BD Biosciences.
Cell viability.
To determine the effect of STAT3 inhibitors, JSI-124 and S3I-201, on the viability of human NK cells, 106 primary NK cells were seeded per well in 24-well plates. JSI-124 or S3I-201 was added at the indicated final concentration (0.05 to 0.5 μM for JSI-124; 5 to 50 μM for S3I-201). At 24, 48, and 72 hours, cells were collected, stained with 7-AAD, and analyzed by flow cytometry.
Flow cytometry analysis.
For direct surface staining, cells were stained with indicated antibodies for 30 minutes at 4°C, washed, and resuspended in staining buffer. Data were acquired using a FACSCalibur cytometer (Becton Dickinson, Franklin Lakes, NJ), and analyzed using FloJo software (Ashland, OR).
Degranulation assay.
Primary NK cells were pretreated with 20 ng/mL of IL-10 or IL-21 with or without 0.1 μM of JSI-124 for 24 hours. Effector and target cells were then cocultured at 1:1 E:T ratio with anti-LAMP1 (CD107a)17 or isotype control antibodies present during the coculture period. Cocultures were incubated for 4 hours at 37°C. After 4 hours incubation, cells were stained with anti-CD56 antibody, and NK-cell degranulation (CD107a expression) was assessed by flow cytometry.
Western blot analysis of STAT phosphorylation.
Primary NK cells were cultured for 24 hours as for degranulation assay, and then lysed with 50 mM Tris-Cl (pH 6.8), 100 mM dithiothreitol, 2% sodium dodecyl sulfate (SDS), and 10% glycerol. Samples were analyzed by SDS-polyacrylamide gel electrophoresis, followed by immunoblotting using Chemo Glow chemiluminescent substrate (Alpha Innotech, San Leandro, CA), according to the manufacturer’s instructions.
STAT3 binding site analysis.
To identify STAT3 binding sites, 10 kb of genomic sequence upstream of the NKG2D translational start site was analyzed using TFSEARCH software (Yutaka Akiyama: “TFSEARCH: Searching Transcription Factor Binding Sites.” http://www.cbrc.jp/research/db/TFSEARCH.html).
Chromatin immunoprecipitation (ChIP) assay.
Primary NK cells were cultured for 24 hours as for degranulation assay, and then ChIP was performed using the STAT3 ChIP Assay Kit (Upstate Biotechnology, Lake Placid, NY). Briefly, equal numbers of primary NK cells were cultured under each condition for 24 hours, fixed with 1% formaldehyde, pelleted, and resuspended in SDS lysis buffer. Chromatin was sonicated to 200∼1000 bp fragments using a Branson Sonifier 250 (Branson Ultrasonics, Danbury CT). Immunoprecipitation was performed using anti-pSTAT3, or mouse IgG as control, and incubated overnight at 4°C with rotation. Immune complexes were collected on protein A-agarose in a solution containing 0.4 μg/μL sonicated salmon sperm DNA, and eluted. Cross-linking was reversed by heating overnight at 65°C. DNA was purified and subsequently amplified by polymerase chain reaction (PCR). A 373-bp fragment from −8828 to −8466, a 181-bp fragment from –4348 to −4168, and a 198-bp fragment from –2977 to −2780 in the upstream of NKG2D translational start site were amplified by PCR using the following primers: forward1 5′-AGA GGA GAC TAC ACC TCA GAC-3′, reverse1 5′-AAG CAA TTC CGA GGC CCT GG-3′; forward2 5′-CTG GCT TAG GAA GCT GTG CC-3′, reverse2 5′-TTC CAT TCA GCT AGG TAT TAA GTA-3′; forward3 5′-CTC AAT TCT GTA TTT AAG AGA CAA-3′, and reverse3 5′-GAT CTA TAT AGA GGA AAT ATA GGC-3′. The PCR product was then analyzed after electrophoresis through 1% agarose gel.
nCounter digital multiplexed gene expression analysis.
Gene expression in NK cells stimulated with mbIL15 or mbIL21 was assessed using the nCounter platform (NanoString Technologies, Seattle, WA). Purified NK cells from 4 donors were stimulated for 3 weeks in parallel with cytokine-expressing aAPCs. Total RNA was isolated from NK cells after initial purification and again after 3 weeks of expansion. Expression of 413 genes was sum-normalized, and selected genes plotted as mean ± standard deviation.
Statistical analysis.
Results are expressed as the mean ± standard deviation. Analysis was performed for the indicated statistical tests using GraphPad Prism for Macintosh, version 5.0a. P values <.05 were considered significant.
Results
IL-21 signaling in NK cells elicits a greater increase in NKG2D expression during ex vivo expansion than IL-15
We had previously shown that propagation of NK cells using aAPCs expressing membrane-bound fusions of IL-15 (mbIL15) or IL-21 (mbIL21) yielded similar percentages of cells expressing NKG2D,15 and that mbIL21 induced strong STAT3 phosphorylation.18 However, we also observed that the level of NKG2D expression was significantly greater in NK cells activated with IL-21 (Figure 1A). To determine whether this was caused by increased transcription of NKG2D, or of its signaling coreceptors DAP10 and DAP12, we assessed the messenger mRNA (mRNA) transcripts for these molecules on freshly-purified human NK cells compared with those expanded with mbIL15 or mbIL21. We found that both conditions caused a similar increase in DAP10 and DAP12 co-receptor transcripts, but that only mbIL21 significantly increased NKG2D transcripts compared with fresh NK cells (Figure 1B).
NK cells expanded ex vivo with aAPCs expressing mbIL21 have higher expression of NKG2D than those expanded with mbIL15. (A) Surface imunophenotyping by flow cytometry was performed on NK cells from each donor after expansion for 3 weeks with the indicated aAPCs. MFI of NKG2D expression is shown for the CD3−CD56+ population. Four donors in this experiment are representative of over 20 donors tested. P values indicated are for 2-tailed Student paired t test. (B) mRNA was purified from freshly-purified NK cells, or NK cells expanded for 3 weeks as above, and assessed for gene expression copy number of DAP10, DAP12, and NKG2D transcripts on the NanoString platform. Five donors in this experiment are representative of 2 independent experiments. P values indicated are for Student t tests of comparisons to fresh NK cells, with Holm-Sídák correction for multiple comparisons. *P < .05; **P < .01; ***P < .001. NS, no statistical significance.
NK cells expanded ex vivo with aAPCs expressing mbIL21 have higher expression of NKG2D than those expanded with mbIL15. (A) Surface imunophenotyping by flow cytometry was performed on NK cells from each donor after expansion for 3 weeks with the indicated aAPCs. MFI of NKG2D expression is shown for the CD3−CD56+ population. Four donors in this experiment are representative of over 20 donors tested. P values indicated are for 2-tailed Student paired t test. (B) mRNA was purified from freshly-purified NK cells, or NK cells expanded for 3 weeks as above, and assessed for gene expression copy number of DAP10, DAP12, and NKG2D transcripts on the NanoString platform. Five donors in this experiment are representative of 2 independent experiments. P values indicated are for Student t tests of comparisons to fresh NK cells, with Holm-Sídák correction for multiple comparisons. *P < .05; **P < .01; ***P < .001. NS, no statistical significance.
STAT3 inhibition decreases NKG2D expression
Since IL-21 signals primarily through STAT3, we then determined whether there was a direct relationship between NKG2D expression and STAT3 activity using the small molecule STAT3 inhibitors, JSI-124 and S3I-201.
To first assess for direct toxicity of STAT3 inhibitors, viability was determined for human NK cells treated with JSI-124 and S3I-201 using 7-AAD staining. STAT3 inhibition did not significantly affect NK cell viability after 24 hours at any concentration, but had some toxicity after 48 and 72 hours (Figure 2A). To minimize confounding effects of direct toxicity, NK cells were treated with lower concentrations of inhibitors for only 24 hours in subsequent functional studies.
STAT3 blockade decreases baseline NKG2D expression of primary human NK cells. (A) To assess the potential direct toxicity of STAT3 inhibitors, freshly-purified human NK cells were cultured in the presence of the STAT3 inhibitors, JSI-124 and S3I-201. NK-cell viability was evaluated at 24, 48, and 72 hours by flow-cytometric analysis of 7-AAD staining. (B) To assess the effect of STAT3 inhibition on NKG2D expression, NK cells were treated with the nontoxic concentrations of STAT3 inhibitors (JSI-124 and S3I-201) for 24 hours, after which NKG2D expression was evaluated by flow cytometry. Replicates are from 3 different donors. P values indicated are for 2-tailed Student t tests of comparisons to untreated NK cells with Bonferroni correction. *P < .05; ***P < .001. NS, no statistical significance.
STAT3 blockade decreases baseline NKG2D expression of primary human NK cells. (A) To assess the potential direct toxicity of STAT3 inhibitors, freshly-purified human NK cells were cultured in the presence of the STAT3 inhibitors, JSI-124 and S3I-201. NK-cell viability was evaluated at 24, 48, and 72 hours by flow-cytometric analysis of 7-AAD staining. (B) To assess the effect of STAT3 inhibition on NKG2D expression, NK cells were treated with the nontoxic concentrations of STAT3 inhibitors (JSI-124 and S3I-201) for 24 hours, after which NKG2D expression was evaluated by flow cytometry. Replicates are from 3 different donors. P values indicated are for 2-tailed Student t tests of comparisons to untreated NK cells with Bonferroni correction. *P < .05; ***P < .001. NS, no statistical significance.
To study the effect of baseline STAT3 activation on NKG2D expression, purified primary human NK cells were incubated with JSI-124 and S3I-201 for 24 hours in the absence of exogenous cytokine. STAT3 inhibition resulted in a significant dose-dependent downregulation of NKG2D surface expression (Figure 2B). Thus, freshly-obtained human NK cells have constitutive STAT3 activation which augments basal NKG2D surface expression.
STAT3 activation increases NKG2D expression
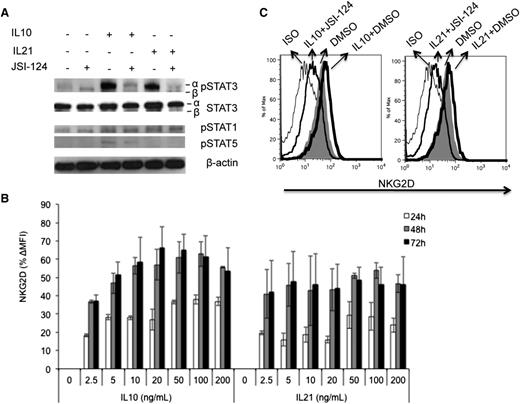
Since inhibition of constitutive STAT3 activation decreased basal NKG2D expression and cytolytic activity, we determined whether additional STAT3 activation affected NKG2D expression and cytolytic activity of NK cells by stimulation with STAT3-activating cytokines, IL-10 and IL-21, with or without the STAT3 inhibitor JSI-124. To verify STAT3 activation, tyrosine705-phosphorylated STAT3 (pSTAT3) was measured by western blot. As expected, IL-10 and IL-21 significantly increased STAT3α phosphorylation and had no effect on STAT3β phosphorylation (Figure 3A), and STAT3α phosphorylation was significantly reduced in the presence of STAT3 inhibitor. Both IL-10 and IL-21 induced minimal phosphorylation of STAT1, and the STAT3 inhibitor did not affect basal or induced pSTAT1 levels at this concentration. IL-10, but not IL-21, also induced slight tyrosine694 phosphorylation of STAT5 (pSTAT5), which was also inhibited by JSI-124 (Figure 3A).
Enhanced NKG2D expression on primary human NK cells by IL-10 and IL-21 is STAT3 dependent. (A) To assess the activation of STAT1, STAT3, and STAT5, NK cells were treated with 20 ng/mL of IL10 or IL21, with or without 0.1 µM of JSI-124 for 24 hours. Total and tyrosine-phosphorylated STAT were evaluated by western blot of cell lysates using STAT- or phospho–STAT-specific antibodies as indicated. (B) To assess cytokine-mediated induction of NKG2D expression, primary NK cells were treated with IL-10 and IL-21 at the indicated concentrations. After treatment of 24, 48, and 72 hours, NKG2D expression was evaluated by flow cytometry. (C) To assess the contribution of STAT3 signaling to cytokine-mediated induction of NKG2D, primary NK cells were treated with 20 ng/mL IL-10 or IL-21 with or without 0.1 µM of JSI-124 for 24 hours. NKG2D surface expression on NK cells was then determined by flow cytometry. (A,C) Representative of at least 3 independent experiments. (B) Pooled data from 4 donors.
Enhanced NKG2D expression on primary human NK cells by IL-10 and IL-21 is STAT3 dependent. (A) To assess the activation of STAT1, STAT3, and STAT5, NK cells were treated with 20 ng/mL of IL10 or IL21, with or without 0.1 µM of JSI-124 for 24 hours. Total and tyrosine-phosphorylated STAT were evaluated by western blot of cell lysates using STAT- or phospho–STAT-specific antibodies as indicated. (B) To assess cytokine-mediated induction of NKG2D expression, primary NK cells were treated with IL-10 and IL-21 at the indicated concentrations. After treatment of 24, 48, and 72 hours, NKG2D expression was evaluated by flow cytometry. (C) To assess the contribution of STAT3 signaling to cytokine-mediated induction of NKG2D, primary NK cells were treated with 20 ng/mL IL-10 or IL-21 with or without 0.1 µM of JSI-124 for 24 hours. NKG2D surface expression on NK cells was then determined by flow cytometry. (A,C) Representative of at least 3 independent experiments. (B) Pooled data from 4 donors.
NKG2D expression showed a substantial increase in expression upon IL-10 and IL-21 stimulation, peaking at 48 hours (Figure 3B). IL-10–mediated NKG2D expression was concentration-dependent, reaching a plateau at 20 ng/mL. The peak expression of NKG2D induced by IL-21 was less than that induced by IL-10, but was achieved at a lower concentration (2.5 ng/mL) and was sustained over a wider range of concentrations (Figure 3B). Activation of NK cells with IL-10 and IL-21 in the presence of JSI-124 not only blocked the cytokine-induced increase in NKG2D, but also decreased NKG2D expression to below basal levels (Figure 3C). These results demonstrate that the increase in NKG2D expression mediated by IL-10 and IL-21 in NK cells is STAT3-dependent.
Next, we evaluated the effect of STAT3-activating cytokines on the degranulation response of NK cells when exposed to K562 tumor targets. CD107a was assessed on NK cells after treatment with IL-10 and IL-21, followed by co-incubation with K562. Both IL-10 and IL-21 (Figure 4) caused a significant increase in degranulation of NK cells in response to K562. Conversely, degranulation was inhibited back to baseline levels when STAT3 phosphorylation was suppressed with JSI-124.
STAT3 activation enhances NK-cell degranulation and cytotoxicity. Primary human NK cells were treated with 20 ng/mL of IL10 or IL21 with or without 0.1 µM of JSI-124 for 24 hours, and followed by coincubation with K562 cells for 4 hours. Target-mediated NK-cell degranulation was assessed by flow cytometry for CD107a. (A) Representative dot plot; (B) Pooled data from 3 donors. P values are shown for 2-tailed unpaired Student t tests.
STAT3 activation enhances NK-cell degranulation and cytotoxicity. Primary human NK cells were treated with 20 ng/mL of IL10 or IL21 with or without 0.1 µM of JSI-124 for 24 hours, and followed by coincubation with K562 cells for 4 hours. Target-mediated NK-cell degranulation was assessed by flow cytometry for CD107a. (A) Representative dot plot; (B) Pooled data from 3 donors. P values are shown for 2-tailed unpaired Student t tests.
Thus, IL-10 and IL-21 increase NKG2D surface expression on human NK cells through a mechanism involving STAT3 phosphorylation, and accordingly, enhance NK-cell degranulation. These results demonstrate a role for STAT3 activation in the regulation of NK-cell responsiveness to target cells.
NKG2D expression is lower in NK cells with conditional STAT3 deletion
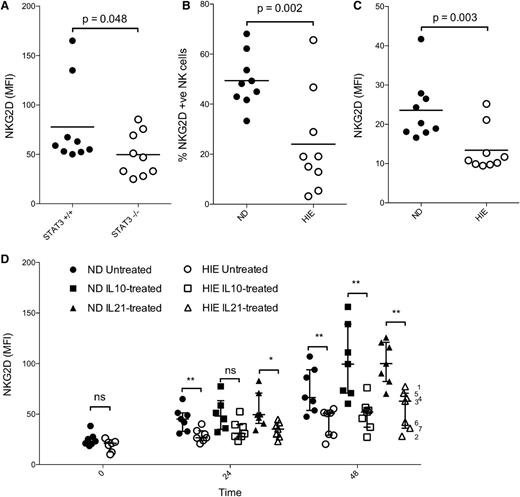
Cytokines are well known to be promiscuous in their activation of signaling pathways, and all STAT inhibitors have some degree of overlapping specificity due to high homology between the SH2 domains on STATs, which they target. Therefore, in a more specific genetic approach, we measured NKG2D expression on NK cells obtained from mice with conditional deletion of floxed STAT3 limited to hematopoietic cells through Tie2 activation of Cre. NK cells obtained from STAT3−/− mice demonstrated a significantly lower expression of NKG2D compared with those obtained from WT mice (Figure 5A), corroborating the pharmacologic evidence for the regulation of NKG2D receptor expression by STAT3 activation.
Conditional ablation of STAT3 in murine hematopoietic cells or congenital STAT3 dysfunction in humans results in decreased basal NKG2D surface expression and decreased upregulation by STAT3-activating cytokines. (A) NK cells were isolated from the spleens of STAT3 conditional knockout and WT mice. The expression of NKG2D on NK1.1+ CD3− splenocytes was determined by flow cytometry. PBMCs were isolated from NDs and patients with HIE and NK cells (gated on CD56+ CD3− lymphocytes), and were assessed for baseline expression of NKG2D by flow cytometry and expressed in panel (B) as MFI, or in panel (C) as % of positive cells compared with isotype controls. (D) PBMCs were then cultured with 20 ng/mL of IL10 or IL21, and NKG2D expression determined at 24 and 48 hours. Data were obtained in 2 separate experiments by different investigators, the results of which were pooled after normalization of the 2 experiments to mean expression at 48 hours. P values for 1-tailed unpaired Student t tests are shown or designated as *P < .05; **P < .01. NS, no statistical significance. HIE sample numbers from Table 1 are indicated at the far right.
Conditional ablation of STAT3 in murine hematopoietic cells or congenital STAT3 dysfunction in humans results in decreased basal NKG2D surface expression and decreased upregulation by STAT3-activating cytokines. (A) NK cells were isolated from the spleens of STAT3 conditional knockout and WT mice. The expression of NKG2D on NK1.1+ CD3− splenocytes was determined by flow cytometry. PBMCs were isolated from NDs and patients with HIE and NK cells (gated on CD56+ CD3− lymphocytes), and were assessed for baseline expression of NKG2D by flow cytometry and expressed in panel (B) as MFI, or in panel (C) as % of positive cells compared with isotype controls. (D) PBMCs were then cultured with 20 ng/mL of IL10 or IL21, and NKG2D expression determined at 24 and 48 hours. Data were obtained in 2 separate experiments by different investigators, the results of which were pooled after normalization of the 2 experiments to mean expression at 48 hours. P values for 1-tailed unpaired Student t tests are shown or designated as *P < .05; **P < .01. NS, no statistical significance. HIE sample numbers from Table 1 are indicated at the far right.
STAT3-induced NKG2D receptor expression is impaired in NK cells from subjects with dominant-negative STAT3 mutations
In a similar manner, we assessed NKG2D receptor expression on NK cells in a human genetic model using peripheral blood obtained from HIES patients carrying STAT3 mutations (Table 1). Like the STAT3-deficient murine NK cells, human NK cells from HIES patients functionally deficient for STAT3 showed a significantly lower basal NKG2D expression on NK cells (Figure 5B), and decreased percentage of NKG2Dpos NK cells (Figure 5C) compared with healthy NDs. Next, we evaluated NKG2D expression after stimulation of PBMCs with IL-10 and IL-21. NK cells from HIES patients showed a significant blunting of cytokine-mediated NKG2D upregulation compared with NDs (Figure 5D).
STAT3 mutations identified in HIES subjects used in this study
| Subject . | Genotype . | Protein . | Region . |
|---|---|---|---|
| 1 | 1144 C-T | R382W | DNA-binding |
| 2 | 1970 A-G | Y657C | SH2 |
| 3 | 1909 G-A | V637M | SH2 |
| 4 | 2003 C-T | S668F | SH2 |
| 5 | 1865 C-T | T622I | SH2 |
| 6 | 1903 C-G | Q635E | SH2 |
| 7 | 1903 C-G | Q635E | SH2 |
| Subject . | Genotype . | Protein . | Region . |
|---|---|---|---|
| 1 | 1144 C-T | R382W | DNA-binding |
| 2 | 1970 A-G | Y657C | SH2 |
| 3 | 1909 G-A | V637M | SH2 |
| 4 | 2003 C-T | S668F | SH2 |
| 5 | 1865 C-T | T622I | SH2 |
| 6 | 1903 C-G | Q635E | SH2 |
| 7 | 1903 C-G | Q635E | SH2 |
Phosphorylated STAT3 binds upstream of NKG2D
As STAT3 mediates its gene-regulating effect by binding to STAT3-specific target sequences, we determined whether STAT3 binds in the region proximal to the NKG2D gene. First, we analyzed the DNA sequence upstream of the NKG2D translational start site using TFSEARCH software, and found 5 potential sites that matched the canonical STAT3 binding sequence (TTCCX(3-4)AA). Three sites were very close to each other, collectively designated as binding site 1, and the 2 remaining sites were designated as binding site 2 and 3, respectively (Figure 6A). To confirm binding of phosphorylated STAT3, primers specific to each site were used to amplify DNA recovered by chromatin immunoprecipitated from NK cells with anti-pSTAT3 mAb. Phosphorylated STAT3 immunoprecipitation recovered DNA from binding site 1, but not 2 and 3 (Figure 6B). This binding was observed in unstimulated NK cells, indicating that constitutive levels of STAT3 activation in resting NK cells is sufficient for nuclear translocation and binding to NKG2D-regulating sites. This binding was further increased by IL-10 and IL-21 stimulation of NK cells, and was inhibited in the presence of JSI-124 (Figure 6C). Taken together, these results demonstrate a direct interaction between activated STAT3 and putative regulatory elements of the NKG2D gene, and suggest direct transcriptional regulation of NKG2D by phosphorylated STAT3.
Phosphorylated STAT3 binds upstream of the human NKG2D gene translational start site. (A) The genomic sequence upstream of the NKG2D start site was analyzed using TFSEARCH software to identify putative STAT3 binding elements. Three potential regions were identified, with STAT3 binding elements indicated in underlined bold font. Sequences flanking the binding elements (identified in underlined font) were used as PCR primers. (B) Constitutive binding of pSTAT3 to the putative STAT3 binding sites was determined by PCR for each predicted region following co-ChIP with phosphorylated STAT3 from nonactivated primary human NK cells. (C) Enhanced binding of pSTAT3 to site 1 was assessed in response to treatment of NK cells with 20 ng/mL of IL10 or IL21, with or without JSI-124 for 24 hours. Data are representative of 2 replicate experiments.
Phosphorylated STAT3 binds upstream of the human NKG2D gene translational start site. (A) The genomic sequence upstream of the NKG2D start site was analyzed using TFSEARCH software to identify putative STAT3 binding elements. Three potential regions were identified, with STAT3 binding elements indicated in underlined bold font. Sequences flanking the binding elements (identified in underlined font) were used as PCR primers. (B) Constitutive binding of pSTAT3 to the putative STAT3 binding sites was determined by PCR for each predicted region following co-ChIP with phosphorylated STAT3 from nonactivated primary human NK cells. (C) Enhanced binding of pSTAT3 to site 1 was assessed in response to treatment of NK cells with 20 ng/mL of IL10 or IL21, with or without JSI-124 for 24 hours. Data are representative of 2 replicate experiments.
Discussion
NKG2D is the primary activating receptor on NK cells responsible for eliciting antitumor cytolytic activity,19 constitutively expressed on resting NK cells and increased upon activation.11 However, the molecular mechanisms regulating NKG2D expression are poorly defined. Here, we used a pharmacologic model to measure NKG2D expression at the protein and mRNA levels in normal human NK cells treated with STAT3 inhibitors, and/or STAT3 activating cytokines and showed that STAT3 activation results in increased NKG2D expression. We then used murine and human genetic models to show that NKG2D expression is decreased on murine STAT3−/− NK cells, and NK cells from patients with dominant-negative STAT3 mutations have decreased NKG2D responses to STAT3-activating cytokines. Lastly, we demonstrated that STAT3 binding to a putative binding site upstream of NKG2D increases or decreases with STAT3 activation and inhibition, respectively. Although cytokines are known to modulate NKG2D expression, to our knowledge these findings are the first to demonstrate transcriptional regulation of NKG2D expression. Whereas immunosuppression mediated by STAT3 activation has been well-described,20 in this study, we show evidence for an immune-activating role for STAT3.
IL-2, IL-7, and IL-15, cytokines belonging to the common γ chain family, are known to enhance expression of NKG2D on NK cells.21 IL-2 and IL-15, which predominantly activate STAT5 in NK cells,22 are associated with moderately decreased mRNA and increased surface protein expression of NKG2D.23 These cytokines modulate NKG2D expression at the posttranscriptional level by increasing transcription and translation of DAP10, thereby enhancing the availability of this adaptor protein that is required for the NKG2D protein expression on the cell surface.23 IL-2, in combination with IL-18, has been shown to prevent transforming growth factor-β–mediated downregulation of NKG2D expression in NK92 cells via c-Jun N-terminal kinase pathway. It is not known whether this modulation is at the transcriptional or translational level,24 though enhanced availability of DAP10 is also likely in this setting, as c-Jun N-terminal kinase activates AP1, which in turn activates the transcription of DAP10.25 IL-12 and IFN-α are also known to induce NKG2D expression,12,26 while transforming growth factor-β1 and prolonged exposure to NKG2D ligands downregulate NKG2D expression.27,28 Silencing of STAT3 in NK92, a human cell line derived from an NK-like large granular lymphoma, resulted in reduced cytotoxicity, and silencing of suppressor of cytokine signaling 3, a negative regulator of STAT3 signaling, led to increased NK92 cytotoxicity,29 though a mechanism for the effect on cytotoxicity was not reported.
In addition to the role of constituitively phosphorylated STAT3 in regulating basal NKG2D expression in nonactivated NK cells, we observed further increases in STAT3 phosphorylation, binding of pSTAT3 to the NKG2D promoter region, and subsequent induction of NKG2D surface expression upon stimulation of NK cells with the STAT3-activating cytokines IL-10 and IL-21. This increase in STAT3 binding and NKG2D expression was reduced in the presence of the STAT3 inhibitor. Given the distance of this binding site from the transcriptional start site (5.2 kb) and the constitutive expression of NKG2D, even in the complete absence of pSTAT3, suggests that the binding site more likely functions as an enhancer than a core promoter. These results are in agreement with previous reports of upregulation of NKG2D on NK cells stimulated with IL-1013 and IL-21,30 though some have reported decreased NKG2D surface expression.31 We focused our investigation on cytokines that predominantly activate STAT3, and did not explore the compound interaction of IL-2 and IL-21 that likely involves coordinated signaling of multiple STAT isoforms. Primary immunodeficiency caused by a loss-of-function mutation in the IL-21 receptor was recently reported in which NK-cell function was severely depressed despite normal NK cell numbers, leading to increased infections and suggesting an important role for STAT3 in NK cell function.32
Because cytokines may exhibit signaling through various secondary pathways, we verified our findings with STAT3-specific pharmacologic and genetic approaches. JSI-124 has been described as a STAT3-specific inhibitor, though it also inhibits the activation of JAK2 and JAK3,33 which indirectly affects activation of other STAT-family members, reinforcing the profound effect we observed on cytotoxicity disproportionate to the effect on NKG2D expression. We also used S3I-201, which binds to the STAT3 SH2-domain with a twofold higher affinity than that of other STAT-family members34 and has no direct JAK inhibition. Binding to the SH2 domain blocks STAT3 recruitment to the cytokine receptor, inhibiting STAT3 phosphorylation and subsequent dimerization, nuclear translocation, and DNA binding.
Genetically engineered mice and NK cells from HIES patients expressing dominant negative STAT3 mutations provided genetic models to test our hypothesis that STAT3 activation regulates NKG2D expression in NK cells. We observed no difference between the basal NKG2D receptor expression on NK cells obtained from HIES patients and those obtained from NDs. This observation suggests that residual STAT3 activity in NK cells from HIES patients is sufficient to maintain constitutive NKG2D expression. However, induction of NKG2D expression mediated through STAT3-activating cytokines was impaired in NK cells from these patients. NK cells play an important early role in immune responses to viral infections, particularly of the herpesviridae family. Although isolated NK-cell deficiency is rare, the early finding of severe herpesvirus infections in a patient with low NK-cell function35 is supported by observations of low NK-cell function in patients with recurrent herpesvirus infections.36 Along with recurrent bacterial infections, HIES patients also have difficulty controlling herpesviridae infections,4 and may have an increased risk of lymphomas.37 Given the role of NK cells in the immunosurveillance of virally-infected and malignant cells through recognition of NKG2D ligands, further investigation of NK-cell dysfunction in HIES patients is warranted.
In this study, we show that NK cells from STAT3-deficient mice have decreased NKG2D expression, which would predict reduced antitumor activity in tumor-bearing models. Kortylewski et al9 demonstrated that ablation of STAT3 led to enhanced rejection of tumors, though this effect was T-cell dependent in an NKG2D–ligand-deficient tumor model. Nonetheless, they demonstrated increased NK-cell cytotoxicity in STAT3−/− tumor-bearing mice. Combined with the data presented here, this suggests that the T-cell activation associated with STAT3 blockade may lead to enhanced NK-cell activity via a secondary STAT3-independent pathway. This is further supported by observations that in vivo antitumor effects of IL-21 are mediated by NK cells and are largely NKG2D dependent.38 We have not addressed whether other STAT-family members also regulate NKG2D, either independent of or as heterodimers with STAT3.
STAT3 inhibition is an attractive anticancer approach for many cancers by virtue of the multifaceted role of STAT3 activation in tumor oncogenesis,39 microenvironment,40 and immunity.41 Tumor-secreted factors such as vascular endothelial growth factor, IL-6, and IL-10 may simultaneously promote tumor growth and immune escape by activating STAT3 signaling in tumor cells, and induce immune suppression by activation of STAT3 in immune cells.42 Likewise, IL-10 is primarily viewed as a negative regulator of immunity,43 but is also known to activate NK-cell antitumor activity in vitro44 and in vivo.45 As NKG2D plays a major role in NK-mediated antitumor activity, the stimulatory effect of IL-10 on NKG2D expression, as observed in this study, may partially explain the antitumor effect of IL-10. It is possible that increased NKG2D expression in NK cells in response to these otherwise anti-inflammatory cytokines, serves as a mechanism to enhance surveillance for stress-related proteins and overcome viral and tumor mechanisms of escaping adaptive immunity.
We previously identified a role for IL-21 in mediating enhanced proliferation and survival of human NK cells through maintenance of telomere length. Further dissection of the effects of STAT-family signaling on NK cells will be important in understanding NK-cell biology and in the application of NK-cell immunotherapy for malignant and infectious diseases. How STAT3 signaling in NK cells affects oncogenesis should be further investigated in in vivo tumor models that discriminate innate and adaptive responses and NKG2D-dependent and -independent mechanisms.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by funding provided by the University of Texas MD Anderson Cancer Center Physician Scientist Program, the St. Baldrick’s Foundation (http://www.stbaldricks.org/), the Farrah Fawcett Foundation, Mr and Mrs David T. Herr, and the National Cancer Institute Cancer Center Support (Core) Grant (CA 16672).
Authorship
Contribution: S.Z. designed and performed the experiments, analyzed data, and wrote the manuscript; P.V.P. designed and performed the experiments, analyzed data, and wrote the manuscript; C.J.D. performed the experiments; V.V.S. performed the experiments and analyzed data; S.S.S. performed the experiments; H.T.N. designed and performed experiments; E.M.M. performed experiments; A.F.F. contributed vital patient samples; S.S.W. contributed vital murine samples; J.S.O. contributed vital patient samples and edited the manuscript; S.M.H. contributed vital patient samples; and D.A.L. designed the research, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for S.Z. is School of Basic Medicine, Shanghai University of Traditional Chinese Medicine, 1200 Cai Lun Rd, Shanghai 201203, China.
Correspondence: Dean A. Lee, MD Anderson Cancer Center, 1515 Holcombe Blvd, Pediatrics Research, Unit #853, Houston, TX 77030; e-mail: dalee@mdanderson.org.
References
Author notes
S.Z. and P.V.P. contributed equally to this study.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal