Key Points
Increased PTK2 expression is associated with improved outcomes in patients with CLL treated with R-FC immunochemotherapy.
PTK2 expression represents a useful, novel biomarker for selection of patients who will benefit from R-FC immunochemotherapy.
Abstract
Addition of rituximab (R) to fludarabine and cyclophosphamide (FC) has significantly improved patient outcomes in chronic lymphocytic leukemia (CLL). Whether baseline gene expression can identify patients who will benefit from immunochemotherapy over chemotherapy alone has not been determined. We assessed genome-wide expression of 300 pretreatment specimens from a subset of 552 patients in REACH, a study of FC or R-FC in relapsed CLL. An independent test set was derived from 282 pretreatment specimens from CLL8, a study of FC or R-FC in treatment-naïve patients. Genes specific for benefit from R-FC were determined by assessing treatment-gene interactions in Cox proportional hazards models. REACH patients with higher pretreatment protein tyrosine kinase 2 (PTK2) messenger RNA levels derived greater benefit from R-FC, with significant improvements in progression-free survival, independent of known prognostic factors in a multivariate model. Examination of PTK2 gene expression in CLL8 patients yielded similar results. Furthermore, PTK2 inhibition blunted R-dependent cell death in vitro. This retrospective analysis from 2 independent trials revealed that increased PTK2 expression is associated with improved outcomes for CLL patients treated with R-FC vs FC. PTK2 expression may be a useful biomarker for patient selection in future trials. These trials were registered at www.clinicaltrials.gov as #NCT00090051 (REACH) and #NCT00281918 (CLL8).
Introduction
Chronic lymphocytic leukemia (CLL) is the most common form of adult leukemia in the Western world. At the molecular level, the disease has substantial heterogeneity and variable clinical outcome.1-3 Nonetheless, the addition of rituximab (R) to chemotherapy has demonstrated high efficacy in CLL4,5 and has significantly prolonged overall survival in untreated CLL and progression-free survival (PFS) in relapsed CLL patients with untreated or relapsed/refractory CLL compared with chemotherapy alone.6-8 Although there is significant benefit of adding R to chemotherapy in multiple hematologic malignancies, the clinical outcome remains variable.
R may exert its antitumor activity by mediating antibody-dependent cellular cytotoxicity (ADCC),9 complement-dependent cytotoxicity (CDC),10 and/or apoptosis via direct signaling.11-13 In CLL, it is not exactly clear which mechanism of action may be the most important for R antitumor activity. Recent evidence has questioned the dominant role of ADCC in the context of immunochemotherapy (eg, R plus fludarabine and cyclophosphamide [R-FC]) and as a monotherapy with R because FC γ-receptor polymorphisms did not influence clinical outcome.14,15 Nonetheless, the variable clinical outcome with R-FC and FC treatment suggests that preexisting biological disease parameters could be important in determining treatment benefit. Indeed, factors such as chromosome 17p deletion (del[17p]), zeta-chain associated protein kinase 70kDa (ZAP70) positivity, high β2-microglobulin, and immunoglobulin heavy chain variable (IGHV) unmutated status remain prognostic indicators of poor clinical outcome even after the introduction of R to the FC regimen.4,5,7,8,15 Whereas prognostic biomarkers help to assess the risk of disease progression irrespective of therapy, predictive biomarkers help to assess the most likely response or outcome to a particular treatment type. However, thus far, no predictive biomarkers have been identified that could select patients that will have a specific benefit of R-FC compared with FC alone. Such biomarkers could offer the option to prospectively manage CLL treatment options, as well as provide a path forward for novel therapeutics that could perhaps target patients showing less clinical benefit.
We performed genome-wide expression profiling on a large number of samples from CLL patients enrolled in a controlled, randomized trial to identify a patient population that would show a prolonged PFS with R-FC therapy compared with FC treatment alone and tested such observations in an independent cohort of patients.
Methods
REACH and CLL8 study design
The REACH study (NCT00090051) was an international, multicenter, open-label, phase 3 study, in which patients with previously treated CLL were randomized (1:1) to receive R-FC (n = 276) or FC alone (n = 276). The CLL8 study (NCT00281918) was also an international, multicenter, open-label, phase 3 study, in which treatment-naïve patients were randomized (1:1) to receive R-FC (n = 408) or FC alone (n = 409). The primary objective of these studies was to demonstrate superior PFS for R-FC compared with FC alone. The study protocols were approved by institutional review boards at participating centers, and all patients gave written informed consent. This study was conducted in accordance with the Declaration of Helsinki. Details on trial design and eligibility criteria and clinical outcome have been described elsewhere.7,8 Patients were selected for gene expression profiling based on the availability of sufficient RNA and gave written informed consent to participate in the additional studies.
Molecular profiling
Pretreatment samples for gene expression profiling analyses were available from 300 of 552 (54%) patients enrolled in the REACH trial and 282 of 817 (35%) from the CLL8 trial. CLL samples were positively enriched by magnetic cell sorting using CD19 microbeads and magnetic-activated cell sorting columns (Miltenyi Biotec Ltd., Surrey, United Kingdom). RNA samples were profiled on the Affymetrix Exon 1.0 ST microarrays. The expression of protein tyrosine kinase 2 (PTK2) was confirmed by real-time quantitative reverse-transcription polymerase chain reaction (qRT-PCR) using an ABI PTK2 kit according to the manufacturer’s protocol and using ubiquitin C as the control gene. All amplifications were performed on a validated ABI 7900HT real-time thermocycler (ABI, Branchburg, NJ).
Statistical analysis
The Exon 1.0 ST microarray probe intensities were background corrected, normalized, and summarized using the robust multiarray averaging method,16 and they included only the core probes of the 17 782 probe sets representing RefSeq genes (Affymetrix annotation version na29.hg18). Only 1244 probe sets with a good dynamic range (fifth to 95th percentiles is at least fourfold and >5% of samples with log2 intensity >5) were subsequently included for outcome analysis. Q values (minimum false discovery rates when tests will be called significant) were calculated to account for multiple testing where appropriate.17
Pretreatment clinical, as well as biological, features and response to therapy among the treatment groups were compared using the Fisher’s exact, Wilcoxon, or log-rank tests. The predictive utility of each gene for the clinical benefit of R-FC was assessed by the statistical significance (Wald test P values) of the treatment-gene interaction term in a Cox proportional hazards model of PFS with treatment (R-FC or FC), gene expression (as a continuous variable), and the treatment-gene interaction term as covariates, as well as by the likelihood ratio (LR) test (with 2 degrees of freedom) comparing the treatment-gene model with the treatment-only model. In addition, to assess whether candidate genes provide predictive or prognostic information independent of known prognostic factors, each gene was also evaluated in the context of an expanded, multivariate Cox proportional hazards model, which included a parsimonious set of known prognostic factors that were statistically significant in both univariate and multivariate models in the overall (intent-to-treat [ITT]) and gene expression study population: age (as a continuous variable), Binet stage (C vs A/B), the Eastern Cooperative Oncology Group (ECOG) performance status (1 vs 0), IGHV mutational status (unmutated vs mutated), and del(17p).15 Additional factors that were considered but were not part of the parsimonious multivariate model included β2-microglobulin (>upper limit of normal vs otherwise), lymphocyte counts (>25 × 109/L vs otherwise), CD38 (overexpression vs otherwise), ZAP70 (overexpression vs otherwise), del(11q), del(13q), and trisomy 12.
All analyses were conducted using the statistical software environment R (http://www.r-project.org).
Microarray data are assigned the following Gene Expression Omnibus accession number: GSE58211.
Cell-line experiments
SU-DHL-6 cells were obtained from the American Type Culture Collection and grown in RPMI 1640 media with 10% fetal bovine serum. Cells were treated with R (Genentech Inc.) and the PTK2 inhibitor (PTK2i), PF-573228 (Sigma), for 24 hours, and cell death was assessed by Annexin V/propidium iodide staining and flow cytometry. Data were normalized to an isotype control antibody. pPTK2 Y397 (Cell Signaling Technologies, Danvers, MA) was used to assess inhibition of PTK2 kinase activity.
Results
The REACH trial gene expression data were available from 148 patients within the FC arm and 152 patients within the R-FC arm. The median follow-up time was 25 months for the gene expression study population, as well as for the overall population.6 The baseline patient demographics and tumor characteristics (supplemental Table 1, available on the Blood Web site) were balanced between the R-FC and FC arms within the gene expression study population. The 2 treatment arms were well balanced when comparing the gene expression study population with the whole study population with respect to risk factors such as age, stage, del(11q), del(17p), IGHV mutational status, and CD38 expression. In the study cohort with available material for gene expression profiling, the treatment benefit with respect to PFS observed in the gene expression study population (median PFS 30.3 months vs 18.5 months, respectively; hazard ratio [HR] = 0.68 [95% confidence interval (CI), 0.49-0.93]; P = .015) was similar to that in the overall population (supplemental Table 2 and supplemental Figure 1), suggesting that the gene expression study population was representative of the REACH overall study population.8
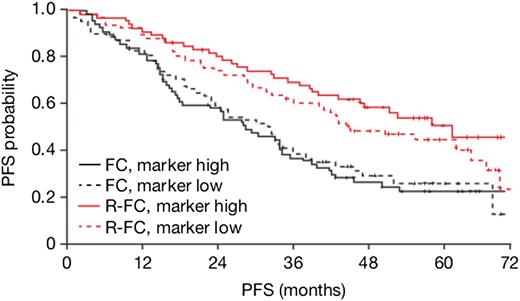
We identified 7 genes indicating different treatment effects dependent on messenger RNA (mRNA) expression levels, 4 of which associate specifically with R-FC, but not FC benefit. These 4 genes included the following: histone cluster 1, H3i (HISTH13I), a member of the histone H3 family; PTK2, a member of the focal adhesion kinases; histone cluster 1, H4k (HIST1H4K), a member of the H4 histone family; and Huntingtin-interacting protein 1 related (HIP1R), encoding for a component of clathrin-coated pits and vesicles, associating specifically with R-FC, but not FC benefit (Table 1). We hypothesized that PTK2 is likely to be a stronger candidate gene for R-FC benefit given that it encodes for a kinase and is involved in B-cell receptor (BCR) cellular signaling.18 In addition, similarities between CD20 and BCR signaling have been described.19 PTK2 mRNA expression level emerged as being predictive for PFS in R-FC (Figure 1 and supplemental Figures 2 and 3), but not FC treatment, regardless of adjusting for known prognostic factors (treatment-PTK2 expression interaction term P = .018/.031 for PTK2 in Cox models without/with known prognostic factors). More specifically, higher expression of PTK2 was correlated with improved clinical outcome as results of treatment with R-FC, but not FC. To confirm that the microarray was capturing PTK2 abundance accurately and not a technical artifact, we performed qRT-PCR on the exact same set of clinical samples and determined whether the data sets were correlated and if the expression of PTK2 by qRT-PCR was also predictive for R-FC vs FC (supplemental Figure 4). Indeed, PTK2 mRNA expression by qRT-PCR was highly correlated with the microarray data (Spearman’s correlation coefficient 0.87) and was predictive of R-FC (treatment-PTK2 expression interaction term P = .02/.03 for PTK2 in Cox models without/with known prognostic factors), but not FC, clinical outcome.
Candidate genes with LR test P < .05 and treatment-mRNA interaction P < .05 in models with or without adjusting for known prognostic factors
| Gene symbol . | Gene name . | Median (5%-95%) . | HR (95% CI) of R-FC vs FC . | P (from expanded model) . | |||||
|---|---|---|---|---|---|---|---|---|---|
| Gene high (>median) . | Gene low (≤median) . | LR test . | Q value of LR test . | Tx-gene interaction . | Gene effect within FC . | Gene effect within R-FC . | |||
| HIST1H3I | Histone cluster 1, H3i | 9.1 (5.4-10.6) | 0.89 (0.59-1.34) | 0.47 (0.28-0.79) | 5e-04 (.00039) | .0098 (.15) | .013 (.029) | .5 (.33) | .00072 (.00043) |
| PTK2 | Protein tyrosine kinase 2 | 5.1 (3.9-7.1) | 0.48 (0.3-0.78) | 0.94 (0.62-1.45) | 5e-04 (.0027) | .0098 (.2) | .018 (.031) | .37 (.69) | .00041 (.00085) |
| HIST1H4K | Histone cluster 1, H4k | 9.3 (7.7-10.3) | 0.79 (0.52-1.19) | 0.55 (0.33-0.91) | .00083 (.00093) | .013 (.15) | .015 (.015) | .67 (.76) | .0017 (.00034) |
| IGF1R | Insulin-like growth factor 1 receptor | 4 (3.3-5.7) | 0.46 (0.3-0.71) | 0.95 (0.59-1.52) | .00096 (.0027) | .014 (.2) | .0017 (.00082) | 6.1e-05 (.0019) | .51 (.017) |
| NR3C2 | Nuclear receptor subfamily 3, group C, member 2 | 4.8 (4.1-6.7) | 0.53 (0.35-0.81) | 0.92 (0.57-1.49) | .0014 (.01) | .016 (.3) | .022 (.019) | .00012 (.00045) | .74 (.4) |
| HIP1R | Huntingtin-interacting protein 1 related | 6.9 (6.1-8.1) | 0.41 (0.25-0.67) | 1 (0.65-1.52) | .013 (.0028) | .052 (.2) | .0064 (.015) | .18 (.86) | .015 (.0012) |
| STAP1 | Signal transducing adaptor family member 1 | 5.2 (4.2-6.3) | 0.61 (0.38-0.96) | 0.73 (0.47-1.14) | .021 (.027) | .068 (.35) | .0059 (.0078) | .032 (.068) | .072 (.082) |
| Gene symbol . | Gene name . | Median (5%-95%) . | HR (95% CI) of R-FC vs FC . | P (from expanded model) . | |||||
|---|---|---|---|---|---|---|---|---|---|
| Gene high (>median) . | Gene low (≤median) . | LR test . | Q value of LR test . | Tx-gene interaction . | Gene effect within FC . | Gene effect within R-FC . | |||
| HIST1H3I | Histone cluster 1, H3i | 9.1 (5.4-10.6) | 0.89 (0.59-1.34) | 0.47 (0.28-0.79) | 5e-04 (.00039) | .0098 (.15) | .013 (.029) | .5 (.33) | .00072 (.00043) |
| PTK2 | Protein tyrosine kinase 2 | 5.1 (3.9-7.1) | 0.48 (0.3-0.78) | 0.94 (0.62-1.45) | 5e-04 (.0027) | .0098 (.2) | .018 (.031) | .37 (.69) | .00041 (.00085) |
| HIST1H4K | Histone cluster 1, H4k | 9.3 (7.7-10.3) | 0.79 (0.52-1.19) | 0.55 (0.33-0.91) | .00083 (.00093) | .013 (.15) | .015 (.015) | .67 (.76) | .0017 (.00034) |
| IGF1R | Insulin-like growth factor 1 receptor | 4 (3.3-5.7) | 0.46 (0.3-0.71) | 0.95 (0.59-1.52) | .00096 (.0027) | .014 (.2) | .0017 (.00082) | 6.1e-05 (.0019) | .51 (.017) |
| NR3C2 | Nuclear receptor subfamily 3, group C, member 2 | 4.8 (4.1-6.7) | 0.53 (0.35-0.81) | 0.92 (0.57-1.49) | .0014 (.01) | .016 (.3) | .022 (.019) | .00012 (.00045) | .74 (.4) |
| HIP1R | Huntingtin-interacting protein 1 related | 6.9 (6.1-8.1) | 0.41 (0.25-0.67) | 1 (0.65-1.52) | .013 (.0028) | .052 (.2) | .0064 (.015) | .18 (.86) | .015 (.0012) |
| STAP1 | Signal transducing adaptor family member 1 | 5.2 (4.2-6.3) | 0.61 (0.38-0.96) | 0.73 (0.47-1.14) | .021 (.027) | .068 (.35) | .0059 (.0078) | .032 (.068) | .072 (.082) |
P values in the parentheses were derived from respective expanded models that included additional known prognostic factors (age, Binet stage, ECOG status, β2-microglobulin, IGHV mutational status, and del[17p]) as covariates, as described in “Methods.” LR test refers to the LR test with 2 degrees of freedom to test for the model with treatment (Tx) + gene + TX-gene interaction vs the treatment-only model, as a way to assess the utility of each mRNA to predict PFS. Gene effect within FC measures the prognostic effect of the gene by computing HRs and associated statistical significance in FC patients with high (≥median) vs low (<median) level of gene expression.
R-FC vs FC effect in terms of PFS in relation to PTK2 expression level (high: >median vs low: ≤median) in the REACH cohort of patients. The curves represent Kaplan-Meier estimates of probability of PFS for REACH patients in treatment (R-FC, red; FC, black) and PTK2 expression level (PTK2 high, solid; PTK2 low, dashed) subgroups.
R-FC vs FC effect in terms of PFS in relation to PTK2 expression level (high: >median vs low: ≤median) in the REACH cohort of patients. The curves represent Kaplan-Meier estimates of probability of PFS for REACH patients in treatment (R-FC, red; FC, black) and PTK2 expression level (PTK2 high, solid; PTK2 low, dashed) subgroups.
The expression of PTK2 was associated with the well-established prognostic factors ZAP70 expression and IGHV mutational status (supplemental Table 3; Wilcoxon’s P = 2E−6, .009, respectively); however, PTK2 was independently significant when incorporating into a multivariate model a parsimonious set of pretreatment factors that demonstrated prognostic significance in the study cohort (treatment FC vs R-FC, age, Binet stage, ECOG status, IGHV mutational status, and del[17p]) (Table 1).
Because PTK2 expression was identified in a retrospective manner in a single cohort of patients from a randomized trial, we wished to test whether this predictive biomarker would have any utility in an independent clinical trial with the same treatment regimen. A total of 282 patient samples with microarray data were available from CLL8 to test this hypothesis. This gene expression subgroup of the CLL8 trial was also comparable with the ITT population of the CLL8 trial with HR = 0.54 for R-FC vs FC (median PFS 55.4 months vs 29.9 months). With regard to prognostic markers, the CLL8 gene expression subgroup was well balanced for IGHV mutational status (R-FC arm 34%, FC arm 35% mutated) and del(17p) (R-FC and FC arm both 8%) and had a higher percentage of del(11q) in the R-FC arm (32% vs 23% in the FC arm). Similar to the results from the REACH cohort, higher PTK2 expression also correlated with an increase in PFS in R-FC arm vs FC (Figure 2 and Table 2; supplemental Figure 5 and supplemental Table 4).
R-FC vs FC effect in terms of PFS in relation to PTK2 expression level (high: >median vs low: ≤median) in the CLL8 cohort of patients. The curves represent Kaplan-Meier estimates of probability of PFS for CLL8 patients in treatment (R-FC, red; FC, black) and PTK2 expression level (PTK2 high, solid; PTK2 low, dashed) subgroups.
R-FC vs FC effect in terms of PFS in relation to PTK2 expression level (high: >median vs low: ≤median) in the CLL8 cohort of patients. The curves represent Kaplan-Meier estimates of probability of PFS for CLL8 patients in treatment (R-FC, red; FC, black) and PTK2 expression level (PTK2 high, solid; PTK2 low, dashed) subgroups.
Summary of REACH and CLL8 clinical outcome with PTK2 expression
| Study description summary . | REACH . | CLL8 . |
|---|---|---|
| R-FC vs FC in second-line CLL . | R-FC vs FC in first-line CLL . | |
| ITT (R-FC vs FC) | HR = 0.65 (30.6 mo vs 20.6 mo), n = 552 (290 PFS events) | HR = 0.56 (51.8 mo vs 32.8 mo), n = 817 |
| Biomarker population with PTK2 gene expression data (R-FC vs FC, PFS) | HR = 0.68 (30.3 mo vs 18.5 mo), n = 300 (156 PFS events) | HR = 0.54 (55.4 mo vs 29.9 mo), n = 282 (172 PFS events) |
| HR of R-FC vs FC in PTK2 low/high subgroups (median cutoff) | 0.94 (20 mo vs 17.9 mo)/0.48 (NA vs 21.5 mo) | 0.6 (45.1 mo vs 32.3 mo)/0.42 (61 mo vs 28.3 mo) |
| HR of PTK2 high vs PTK2 low (median cutoff) in FC/R-FC | 0.86/0.46 | 1.08/0.75 |
| Study description summary . | REACH . | CLL8 . |
|---|---|---|
| R-FC vs FC in second-line CLL . | R-FC vs FC in first-line CLL . | |
| ITT (R-FC vs FC) | HR = 0.65 (30.6 mo vs 20.6 mo), n = 552 (290 PFS events) | HR = 0.56 (51.8 mo vs 32.8 mo), n = 817 |
| Biomarker population with PTK2 gene expression data (R-FC vs FC, PFS) | HR = 0.68 (30.3 mo vs 18.5 mo), n = 300 (156 PFS events) | HR = 0.54 (55.4 mo vs 29.9 mo), n = 282 (172 PFS events) |
| HR of R-FC vs FC in PTK2 low/high subgroups (median cutoff) | 0.94 (20 mo vs 17.9 mo)/0.48 (NA vs 21.5 mo) | 0.6 (45.1 mo vs 32.3 mo)/0.42 (61 mo vs 28.3 mo) |
| HR of PTK2 high vs PTK2 low (median cutoff) in FC/R-FC | 0.86/0.46 | 1.08/0.75 |
NA, not assessed.
PTK2 has typically been described as a key regulator of cell adhesion, proliferation, and migration18 and has been reported to be regulated by the BCR.19 Although an association or direct mechanistic connection with R efficacy has not been previously described, the fact that high expression of PTK2 correlates with improved PFS to R-FC, but not FC, in 2 independent data sets suggests that PTK2 might be a contributor to R-dependent cell death. To test this hypothesis, we assessed if inhibition of PTK2 kinase activity would have any consequence on R-dependent cell death in vitro. SU-DHL-6 cells were pretreated with a PTK2i (PF-573228) or dimethylsulfoxide control for 1 hour and subsequently treated with R for 24 hours prior to assessing cell death (Figure 3A). In this experimental system, inhibition of PTK2 kinase activity by the PTK2i resulted in a 37% reduction of R-dependent cell death (P < .01). Inhibition of PTK2 kinase activity was confirmed by assessing the autophosphorylation site Y397 (Figure 3B). These data suggest that the expression and kinase activity of PTK2 might influence the outcome of R-FC treated patients by directly influencing R activity.
Inhibition of PTK2 blunts R-dependent cell death. (A) SU-DHL-6 cells were treated with R and a PTK2 small-molecule inhibitor (PF-573228) or vehicle control (DMSO) for 24 hours, and cell death was assessed by Annexin V/propidium iodide staining and flow cytometry. Data were normalized to isotype control antibody. (B) Confirmation of pPTK2 Y397 reduction by the PTK2i was determined by western blotting (WB). DMSO, dimethylsulfoxide.
Inhibition of PTK2 blunts R-dependent cell death. (A) SU-DHL-6 cells were treated with R and a PTK2 small-molecule inhibitor (PF-573228) or vehicle control (DMSO) for 24 hours, and cell death was assessed by Annexin V/propidium iodide staining and flow cytometry. Data were normalized to isotype control antibody. (B) Confirmation of pPTK2 Y397 reduction by the PTK2i was determined by western blotting (WB). DMSO, dimethylsulfoxide.
Discussion
Personalized health care in oncology has different transformational potential for patient outcome. The most pertinent examples of this include trastuzumab treatment of human epidermal growth factor receptor 2–positive breast cancer,20 imatinib for BCR–Abelson murine leukemia viral oncogene homolog 1 (ABL1)–positive chronic myeloid leukemia,21 and more recently, vemurafenib for v-raf murine sarcoma viral oncogene homolog (BRAF)–mutant melanoma.22 These successes have largely been attributed to the molecular rationale of the development of these drugs, which postulate that these tumors are “addicted” to a specific oncogenic event.23 Trastuzumab, imatinib, and vemurafenib were designed to deprive the tumor of the substance of this “addiction” and consequently result in tumor shrinkage and clinical benefit. In contrast, R was designed to broadly deplete CD20-positive B cells, which has transformed patient treatment of non-Hodgkin lymphoma and CLL, rather than targeting a specific molecular addiction often found in these diseases, such as amplification of the HER2 gene in breast cancer.1,2,24 Despite the significant benefit of adding R to chemotherapy in multiple hematologic malignancies, for patients with relapsed/refractory CLL, there remains a highly variable clinical outcome, suggesting that preexisting biological determinants of the disease exist, which could predict clinical outcome. The complexity by which R exerts its anticancer activity via multiple mechanisms of action25,26 posed a significant challenge to efforts that aimed to identify a specific population in the context of R treatment.
Preclinical studies have elegantly shown that CDC-mediated activity of R is largely ineffective in CLL because of lower levels of CD20, and that CLL cells may also be susceptible to CD20 shaving following treatment, thereby lowering the pool of available CD20 to mediate CDC.27,28 Therefore, it is conceivable that direct signaling, rather than ADCC14,15 or CDC, may be playing a more dominant role for the observed clinical efficacy. Genome-wide expression profiling has offered the opportunity to identify potential patient subpopulations that would gain maximal benefit of R treatment combined with cyclophosphamide, doxorubicin, vincristine, and prednisone therapy in diffuse large B-cell lymphoma,29 although the identified gene signature is prognostic rather than predictive of added R benefit. Nonetheless, this has not been investigated in CLL in the context of R-FC treatment. Our study takes this a step further by identifying a CLL subpopulation that gains specific benefit of R-FC over FC by expression of PTK2 mRNA.
Intriguingly, NOTCH1 mutations are concurrently found with trisomy 12 in ∼28% of cases30 and have been reported to be negatively associated with R-FC benefit over FC in the CLL8 cohort.31 We did not assess NOTCH1 mutations in the REACH cohort; however, PTK2 effects were robust in the multivariate models, with or without trisomy 12. More specifically, treatment by PTK2 interaction P = .022 with addition of trisomy 12 vs P = .027 in the parsimonious model presented.
PTK2 has long been an attractive drug target for solid tumors given its association with poor clinical outcome and key role in positively regulating cell migration, proliferation, and adhesion by integrating with the integrin signaling network.18 The role of PTK2 in the hematologic malignancies has largely been unexplored, and no specific association with CLL or R has previously been described. It has been suggested that R can elicit cell death in part by homotypic adhesion,32 and that PTK2 is autophosphorylated after engagement of homotypic adhesion by other homotypic adhesion-inducing antibodies. It is therefore conceivable that increased PTK2 expression may enhance R-dependent cell death by this mechanism. This could potentially explain why inhibition of PTK2 kinase activity blunts R-dependent cell death and the association of increased PTK2 expression the specific benefit provided to R-FC in this study. In addition, similarities between CD20 and BCR signaling have been described previously19 and these similarities suggest that PTK2 may play a direct role in mediating CD20 signaling. Further studies investigating the mechanism by which PTK2 contributes to R-dependent cell death should be considered.
In summary, our findings pave the way to potentially identify, a priori, a CLL patient subpopulation that will likely benefit particularly from R-FC treatment, as well as identify a subpopulation that will gain less benefit from R-FC vs FC. This not only creates an opportunity to determine patient outcome prior to R-FC treatment and may enable clinicians to select patients for retreatment with R-FC after relapse, but also identifies a poor prognosis subgroup of patients to test with novel targeted therapies. Given the advent of second-generation anti-CD20 antibodies and novel drugs with alternative mechanisms of action that are in development for CLL, a molecular subpopulation, as presented here, may be of increasing importance. Further studies are warranted to confirm these findings prospectively and assess the most appropriate cutoff of PTK2 expression, as well as confirm the potential predictive role of PTK2 expression for R-based immunochemotherapy.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Ting Wang, Helen Smith, Lindsay Brady, Jamieson Sheffield, and the sample management and operations teams for their efforts in making this study possible. The authors also thank the investigators and patients involved in the REACH and CLL8 trials, as well as Boris V. Afanasiev, of Dorevitch Pathology Laboratory, Frankston Hospital in Frankston, Australia. REACH and CLL8 were sponsored by F. Hoffman‒La Roche Ltd. Third-party medical editing assistance was provided by F. Hoffman‒La Roche Ltd.
Authorship
Contribution: D.D., M.W., N.V., and M.K.W. wrote the manuscript and designed the study; R.-F.Y., G.P., and G.D.-N. performed the statistical analyses and had access to the clinical data; T.Q.N. was involved in the mRNA profiling; A.D. was responsible for the molecular and genetic data generated within the REACH trial; K.F., A.-M.F., M.H., J.B., R.B., A.B., H.D., and S.S. designed and performed the CLL8 study and provided samples and analyzed data within the CLL8 study; S.Y.S. and N.Y. performed and designed the in vitro studies; and X.S., T.R., G.N.S., A.D., P.S.-C., K.W., J.L., J.C., L.L., V.A.R., I.B.-B., C.H.G., M.M., and S.S. were clinical investigators as part of REACH and submitted samples for the study and analyzed data.
Conflict-of-interest disclosure: N.V., N.Y., and R.-F.Y. are employees of Genentech Inc. G.P., T.Q.N., G.D.-N., M.K.W., and M.W. are employees of Roche. The remaining authors declare no competing financial interests.
Correspondence: David Dornan, Gilead Sciences Inc., 333 Lakeside Dr, Foster City, CA, 94404; e-mail: david.dornan@gilead.com.