Key Points
HFE increases Smad1/5/8 phosphorylation and hepcidin expression, and inhibition of BMP signaling abolishes HFE-induced hepcidin expression.
HFE interacts with ALK3, inhibits ALK3 ubiquitination-proteasomal degradation, and increases ALK3 cell-surface expression.
Abstract
Mutations in HFE are the most common cause of hereditary hemochromatosis (HH). HFE mutations result in reduced expression of hepcidin, a hepatic hormone, which negatively regulates iron absorption from the duodenum and iron release from macrophages. However, the mechanism by which HFE regulates hepcidin expression in hepatocytes is not well understood. It is known that the bone morphogenetic protein (BMP) pathway plays a central role in controlling hepcidin expression in the liver. Here we show that HFE overexpression increased Smad1/5/8 phosphorylation and hepcidin expression, whereas inhibition of BMP signaling abolished HFE-induced hepcidin expression in Hep3B cells. HFE was found to associate with ALK3, inhibiting ALK3 ubiquitination and proteasomal degradation and increasing ALK3 protein expression and accumulation on the cell surface. The 2 HFE mutants associated with HH, HFE C282Y and HFE H63D, regulated ALK3 protein ubiquitination and trafficking differently, but both failed to increase ALK3 cell-surface expression. Deletion of Hfe in mice resulted in a decrease in hepatic ALK3 protein expression. Our results provide evidence that HFE induces hepcidin expression via the BMP pathway: HFE interacts with ALK3 to stabilize ALK3 protein and increase ALK3 expression at the cell surface.
Introduction
Hereditary hemochromatosis (HH), an inherited disorder of iron metabolism, is characterized by excess iron absorption from the gut and excess iron release from macrophages, with the potential for multiple organ damage. Mutations in several genes including HFE, TFR2 (encoding transferrin receptor 2), HFE2 (encoding hemojuvelin [HJV]), HAMP (encoding hepcidin), and FPN (encoding ferroportin) can result in HH, with HFE mutations representing the most frequent form of HH.1-3
Hepcidin, a small peptide secreted mainly by the liver, is the central regulator of systematic iron metabolism. Hepcidin negatively regulates iron in circulation by inhibiting iron absorption from the duodenum, iron recycling from the monocyte/macrophage system, and iron mobilization from hepatic stores. Hepcidin inhibits the iron efflux by binding to the sole iron exporter ferroportin and inducing its internalization and degradation.4
We and others have demonstrated that the bone morphogenetic protein (BMP) signaling pathway is critically involved in regulation of hepcidin expression in the liver.5-9 BMPs signal through type II (BMPRII, ACTRIIA, and ACTRIIB) and type I (ALK2, ALK3, and ALK6) serine threonine kinase receptors, and intracellular Smad1/5/8 proteins to regulate transcription of target genes.10 BMPs have been shown to stimulate hepcidin expression in hepatocytes in culture.6,7,11 BMP2 or BMP6 increases hepcidin expression and decreases serum iron levels in mice.8,12 Conversely, genetic deletion of BMP6,8,9 hepatic ALK3,13 HJV,6 or Smad4,5 or administration of BMP ligand antagonists HJV.Fc,12 ALK3-Fc, or BMP type I receptor inhibitor LDN-193189,14 each leads to low hepcidin expression in the liver in mice. All of these data suggest that BMP signaling is an important regulatory pathway for hepcidin expression in hepatocytes.
Previous studies have clearly demonstrated that HJV acts through the BMP pathway to regulate hepcidin expression in the liver.6,7 However, the mechanisms by which HFE and TFR2 regulate hepcidin expression are not well understood. Studies from mouse models suggest that HFE and TFR2 may function in the same pathway, and HFE is limiting in regulating hepcidin expression.15-19 Biochemical studies demonstrate that HFE interacts with transferrin receptor 1 (TFR1) and TFR2, thus regulating hepcidin expression.20-23 However, recent studies show that transgenic HFE-dependent induction of hepcidin in mice does not require TFR224 and that there is no required interaction between HFE and TFR2.24,25 All of these findings suggest independent roles of HFE and TfR2 in regulating hepcidin. Nevertheless, the downstream pathways that mediate HFE activity remain elusive. Interestingly, in Hfe knockout mice and in patients with HFE mutations, phosphorylated Smad1/5/8 and Id1 levels relative to BMP6 or iron levels in livers are reduced compared with controls.16,19,26-29 This suggests that HFE may regulate hepcidin expression through the BMP pathway in liver cells. However, the mechanism by which HFE interacts with the BMP pathway to regulate hepcidin expression remains to be defined.
Here, we provide direct evidence that HFE induces hepcidin expression through the BMP pathway in Hep3B cells. HFE interacts with ALK3 to stabilize ALK3 protein and increase ALK3 cell-surface expression. The 2 HFE mutants associated with HH, HFE C282Y and HFE H63D, regulate ALK3 protein degradation and trafficking differently, but both fail to increase ALK3 cell-surface expression and hepcidin expression.
Methods
Animals
All animal studies were approved by the Institutional Animal Care and Use Committee of Zhejing University. Hfe−/− mice with a C57/Bl/6-129/Ola genetic background were donated by Dr Nancy C. Andrews. All animals were fed with commercial diet and water ad libitum. Liver samples were collected from Hfe−/− mice and wild-type (WT) mice on the same background at 8 to 10 weeks of age.
Breeding and genotyping of mice with a liver-specific deletion of Alk3 (Alk3fl/fl; Alb-Cre) and mice carrying the Hfe∆CD-Myc transgene were performed as previously described.13,18,24 Liver samples were collected from Alk3fl/fl, Alb-Cre and Alk3fl/fl female mice at 8 to 10 weeks of age; and from Hfe∆CD-Myc animals and genetically identical WT male mice at 8 weeks of age.
Cell culture and transfection
COS-7 cells were cultured in DMEM medium (Invitrogen) supplemented with 10% fetal bovine serum. HepG2 and Hep3B human hepatoma cells were cultured in MEM α medium (Invitrogen) supplemented with 10% adult horse serum (Invitrogen). HFE-HA, HFE-Myc, ALK3-HA, ALK3-Flag, ACTRIIA-Myc, TFR2-Myc, or a combination of these plasmids was transiently transfected in 12-well or 24-well plates using FuGENE HD Transfection Reagent (Promega) according to the manufacturer’s instructions. Forty-eight hours after transfection, cell lysates were collected for western blotting, immunoprecipitation, or real-time polymerase chain reaction (PCR) analysis.
Immunoprecipitation and western blotting
COS-7, HepG2, or Hep3B cells were lysed as described previously.30 Cell lysates were incubated with anti-HA (ab18181; Abcam), anti-Myc (sc-40; Santa Cruz Biotechnology), or anti-Flag (F1804; Sigma-Aldrich) antibodies as indicated in different experiments at 4°C overnight. The solutions were then incubated for 5 hours at 4°C with protein A beads (Pierce Biotechnology). Liver lysates (2 mg) were preabsorbed with 60 μL protein G beads for 5 hours. Preabsorbed lysates were incubated with 5 μg rabbit anti-Myc (ab9106; Abcam) at 4°C overnight. Protein A beads were added, and lysates were incubated for 5 hours. Eluted proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis and subjected to western blot analysis under reducing conditions using anti-HA, anti-Myc, or anti-Flag antibodies as indicated.
Endogenous ALK3 protein in Hep3B cells and in liver lysates was examined by western blotting using an ALK3 antibody from Millipore (ABD51).
Protein degradation assay
Hep3B cells were transfected with ALK3-HA in the presence or absence of HFE-Myc. Forty-six hours after the transfection, cells were incubated with cycloheximide (Chx; 100 μg/mL) at 37°C for 0, 1, 2, or 4 hours. Cells were then harvested and subjected to western blot analysis for ALK3 protein levels using the anti-HA antibody.
Protein degradation pathway analysis
For treatment with proteasome and lysosome inhibitors, Hep3B cells were transfected with empty vector or ALK3-Flag (200 ng/mL). Forty-six hours after transfection, cells were treated with increasing amounts of MG132 (0, 5, and 10 μM) or NH4Cl (0, 1, and 10 mM) for 5 hours before the collection of whole-cell lysates for western blot analysis for ALK3 protein expression.
For ubiquitin-mediated ALK3 degradation assays, Hep3B cells were transfected with ALK3-Flag (400 ng/mL) in the presence or absence of HA-ubiquitin (100 ng/mL), with and without HFE-Myc plasmid (50 ng/mL). Forty-eight hours after transfection, cells were lysed for western blot analysis for ALK3 protein expression.
For ubiquitination assay, Hep3B cells were transfected with ALK3-Flag in the presence or absence of HA-ubiquitin or Myc-ubiquitin, with and without HFE-Myc, HFE-HA WT, HFE-HA C282Y, or HFE-HA H63D. Forty-six hours after transfection, cells were treated with MG132 (5 μM) for 5 hours before harvesting. Cell lysates were immunoprecipitated using anti-Flag antibodies and analyzed by western blotting using anti-HA or anti-Myc antibodies.
Cell-surface protein biotinylation
Hep3B cells were transfected with ALK3-Flag in the presence of HFE-HA WT, HFE-HA C282Y, or HFE-HA H63D. Forty-eight hours after transfection, cell-surface proteins were isolated by whole-cell biotinylation and Streptavidin agarose precipitation using Sulfo-NHS-LC-LC-Biotin (Thermo Scientific) according to the manufacturer’s instructions. Negative controls for nonspecific binding of ALK3 and HFE to agarose beads were included by omitting Sulfo-NHS-LC-LC-Biotin. The precipitates were analyzed by western blot for ALK3 and HFE expression on the cell surface. A nonspecific membrane protein was used as a loading control.
Immunofluorescence assays
Hep3B cells were transfected with ALK3-Flag in the absence or presence of HFE-HA WT, HFE-HA C282Y, or HFE-HA H63D. Forty-eight hours after transfection, cells were fixed in 4% paraformaldehyde for 20 minutes and permeabilized in a phosphate-buffered saline solution containing 0.1% Triton X-100 for 4 minutes at room temperature. Cells were then incubated with mouse anti-Flag (Sigma-Aldrich) and rabbit anti-HA (Santa Cruz Biotechnology) antibodies, and followed by incubation with Alexa Fluor 488 donkey anti-mouse IgG and Alexa Fluor 555 donkey anti-rabbit IgG. Images were acquired using a confocal microscope (LEICA SP5).
RNA isolation and real-time PCR analysis
Total RNA was isolated from Hep3B and mouse livers using the Pure LinkTM RNA mini kit (Ambion). First-strand cDNA synthesis was performed using the PrimeScript RT reagent Kit (TAKARA) and was amplified using the ABI Prism 7900 Sequence Detection System (PE Biosystems) with the hepcidin and RPL19 primers as previously described.7 RPL19 was used as an internal control.
Statistical analysis
All data are represented as mean ± standard deviation (SD) of independent replicates (n ≥ 3). Student t test was applied for statistical analysis. A P value of ≤.05 was considered statistically significant.
Results
HFE regulates hepcidin expression through the BMP pathway
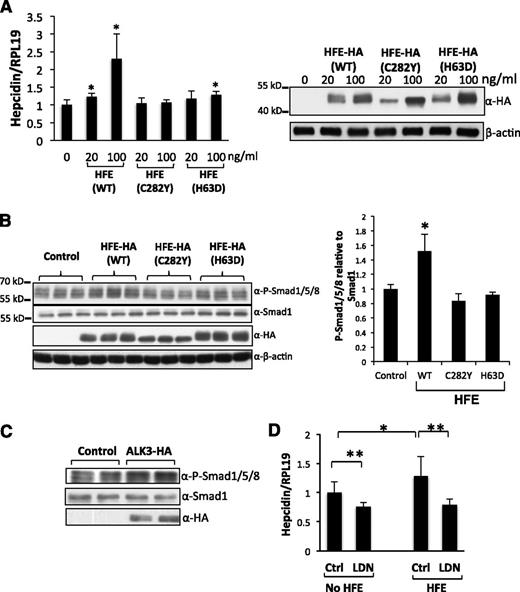
Although it has been known for many years that HFE is an upstream regulator of hepcidin in vivo, this regulation has not been demonstrated in vitro. Therefore, we first examined the effect of overexpression of HFE on BMP signaling and hepcidin expression in Hep3B cells. As shown in Figure 1A, HFE overexpression increased hepcidin expression in a dose-dependent manner. HFE also increased Smad1/5/8 phosphorylation (Figure 1B). As a positive control, transfection of the BMP type I receptor ALK3 increased Smad1/5/8 phosphorylation levels (Figure 1C). Treatment of Hep3B cells with LDN-193189, a small-molecule derivative of the BMP inhibitor dorsomorphin, suppressed both basal and HFE-induced hepcidin expression to similar levels (Figure 1D; compare bars 2 and 4).
HFE increases hepcidin expression through the BMP pathway. (A) Hep3B cells were transfected with WT, HFE-C282Y, or HFE-H63D at 0, 20, and 100 ng/mL. Forty-eight hours after transfection, cells were harvested for real-time PCR analysis for hepcidin mRNA levels, and for western blotting for HFE protein levels. (B) Hep3B cells were transfected with WT, HFE (C282Y), or HFE (H63D) at 100 ng/mL. Cells were harvested for western blotting for phospho-Smad1/5/8, Smad1, and HFE expression (right panel). Phospho-Smad1/5/8 levels obtained by western blot analysis were quantified and normalized to the signal obtained for Smad1 (left panel). (C) Hep3B cells were transfected with empty vector (control) or ALK3-HA expression plasmid. Cells were harvested for western blot analysis for phospho-Smad1/5/8, Smad1, and ALK3 protein expression. (D) Hep3B cells were transfected with or without HFE-Myc (100 ng/mL). Forty-two hours after transfection, cells were treated with or without LDN-193189 (20 nM) for 6 hours. Cells were then collected for real-time PCR analysis for hepcidin and RPL19 mRNA levels. Data represent averages of the values from 3 separate experiments. RPL19 is the internal control. β-actin is the loading control in western blotting. *P < .05, **P < .01 vs the control (bar 1). Data are represented as mean ± SD (n = 3).
HFE increases hepcidin expression through the BMP pathway. (A) Hep3B cells were transfected with WT, HFE-C282Y, or HFE-H63D at 0, 20, and 100 ng/mL. Forty-eight hours after transfection, cells were harvested for real-time PCR analysis for hepcidin mRNA levels, and for western blotting for HFE protein levels. (B) Hep3B cells were transfected with WT, HFE (C282Y), or HFE (H63D) at 100 ng/mL. Cells were harvested for western blotting for phospho-Smad1/5/8, Smad1, and HFE expression (right panel). Phospho-Smad1/5/8 levels obtained by western blot analysis were quantified and normalized to the signal obtained for Smad1 (left panel). (C) Hep3B cells were transfected with empty vector (control) or ALK3-HA expression plasmid. Cells were harvested for western blot analysis for phospho-Smad1/5/8, Smad1, and ALK3 protein expression. (D) Hep3B cells were transfected with or without HFE-Myc (100 ng/mL). Forty-two hours after transfection, cells were treated with or without LDN-193189 (20 nM) for 6 hours. Cells were then collected for real-time PCR analysis for hepcidin and RPL19 mRNA levels. Data represent averages of the values from 3 separate experiments. RPL19 is the internal control. β-actin is the loading control in western blotting. *P < .05, **P < .01 vs the control (bar 1). Data are represented as mean ± SD (n = 3).
There are 2 major mutants for HFE in human patients (ie, HFE-C282Y and HFE-H63D). HFE-C282Y is unable to be expressed on the cell membrane because of the lack of a disulfide bond and the inability to bind β2-microglobulin. HFE-H63D mutant can be expressed on the cell membrane, but it cannot regulate hepcidin.31-33 Therefore, the 2 mutants allow us to investigate the mechanisms by which HFE regulates BMP signaling and hepcidin expression. Transfection of cells with the mutants had no (HFE-C282Y) or little (HFE-H63D) effect on hepcidin expression, although mutant HFE proteins were expressed at levels similar to HFE-WT (Figure 1A). Accordingly, HFE-C282Y or HFE-H63D had no effects on phosphorylated Smad1/5/8 levels (Figure 1B). Together, these results suggest that HFE may upregulate hepcidin via the BMP/Smad pathway.
HFE interacts with ALK3 but not ACTRIIA
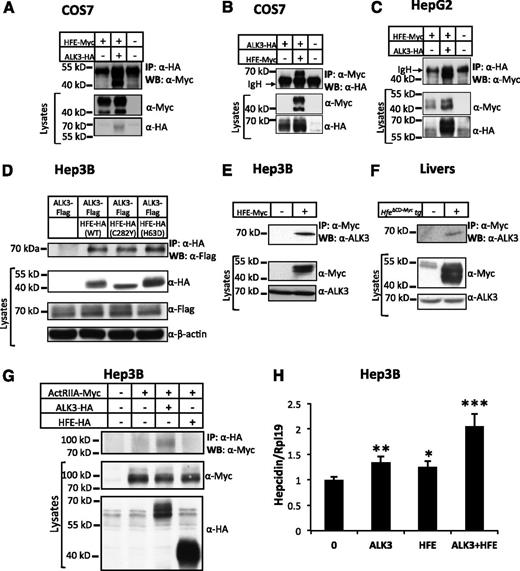
To understand the underlying mechanisms that integrate HFE into the BMP pathway, we examined whether HFE interacts with the BMP receptors by coimmunoprecipitation in COS7, HepG2, and Hep3B cells. We focused on the BMP type I receptor ALK3, because our previous studies suggest that ALK3 plays the most important role among the BMP type I receptors in hepcidin regulation.7,13 We transfected COS7 and HepG2 cells with Myc-tagged HFE (HFE-Myc) in the absence or presence of HA-tagged ALK3 (ALK3-HA). Lysates from transfected cells were subjected to immunoprecipitation of ALK3-HA, and the immunocomplexes were analyzed by western blot analysis for HFE-Myc (Figure 2A,C). HFE-Myc was detected in the HA antibody precipitate from cells transfected with HFE-Myc and ALK3-HA, whereas there were no HFE-Myc bands in the HA antibody precipitates from cells transfected with HFE-Myc alone (Figure 2A,C; compare lanes 2 and 1). In reciprocal anti-Myc immunoprecipitations, ALK3-HA was coimmunoprecipitated with HFE-Myc in COS7 cells (Figure 2B). HFE was also coimmunoprecipitated with ALK3 in Hep3B cells when HFE-HA and ALK3-Flag plasmids were used (Figure 2D; compare lanes 2 and 1). In addition, HFE-Myc was coimmunoprecipitated with ALK6-HA (supplemental Figure 1, available on the Blood Web site).
HFE interacts with ALK3 and synergizes with ALK3 to stimulate hepcidin expression. (A) COS7 cells were transfected with HFE-Myc in the presence or absence of ALK3-HA. Cell lysates were used for immunoprecipitation (IP) and subsequent western blot analysis (WB) using indicated antibodies. Cell lysates were used for WB as indicated to show inputs. (B) COS7 cells were transfected with ALK3-HA in the presence and absence of HFE-Myc. Cell lysates were used for IP and /or WB as indicated. (C) HepG2 cells were transfected with HFE-Myc in the presence or absence of ALK3-HA. Cell lysates were used for IP and /or WB as indicated. (D) Hep3B cells were transfected with ALK3-Flag in the presence of empty vector, HFE-HA (WT), HFE-HA (C282Y), or HFE-HA (H63D). Cell lysates were used for IP and /or WB as indicated. (E) Hep3B cells were transfected with or without HFE-Myc. Cell lysates were used for IP and /or WB as indicated. (F) Lysates from WT and Hfe∆CD-Myc transgenic mice were used for IP and/or WB as indicated. (G) Hep3B cells were transfected with ACTRIIA-Myc in the presence of empty vector, ALK3-HA, or HFE-HA. Cell lysates were used for IP and /or WB as indicated. (H) Hep3B cells were transfected with empty vector (0), ALK3-HA alone, HFE-Myc (20 ng/mL) alone, or ALK3-HA in combination with HFE-Myc expression plasmids. Forty-eight hours after transfection, cells were harvested for real-time PCR analysis for hepcidin and RPL19 mRNA levels. RPL19 is the internal control. *P < .05, **P < .01, ***P < .001. Data are represented as mean ± SD (n = 3 for panel F). All experiments were repeated 2 to 3 times.
HFE interacts with ALK3 and synergizes with ALK3 to stimulate hepcidin expression. (A) COS7 cells were transfected with HFE-Myc in the presence or absence of ALK3-HA. Cell lysates were used for immunoprecipitation (IP) and subsequent western blot analysis (WB) using indicated antibodies. Cell lysates were used for WB as indicated to show inputs. (B) COS7 cells were transfected with ALK3-HA in the presence and absence of HFE-Myc. Cell lysates were used for IP and /or WB as indicated. (C) HepG2 cells were transfected with HFE-Myc in the presence or absence of ALK3-HA. Cell lysates were used for IP and /or WB as indicated. (D) Hep3B cells were transfected with ALK3-Flag in the presence of empty vector, HFE-HA (WT), HFE-HA (C282Y), or HFE-HA (H63D). Cell lysates were used for IP and /or WB as indicated. (E) Hep3B cells were transfected with or without HFE-Myc. Cell lysates were used for IP and /or WB as indicated. (F) Lysates from WT and Hfe∆CD-Myc transgenic mice were used for IP and/or WB as indicated. (G) Hep3B cells were transfected with ACTRIIA-Myc in the presence of empty vector, ALK3-HA, or HFE-HA. Cell lysates were used for IP and /or WB as indicated. (H) Hep3B cells were transfected with empty vector (0), ALK3-HA alone, HFE-Myc (20 ng/mL) alone, or ALK3-HA in combination with HFE-Myc expression plasmids. Forty-eight hours after transfection, cells were harvested for real-time PCR analysis for hepcidin and RPL19 mRNA levels. RPL19 is the internal control. *P < .05, **P < .01, ***P < .001. Data are represented as mean ± SD (n = 3 for panel F). All experiments were repeated 2 to 3 times.
We also examined whether HFE interacts with endogenous ALK3. As shown in Figure 2E, HFE-Myc was co-immunoprecipitated with endogenous ALK3 in Hep3B cells. To detect the interaction in livers, we used transgenic mice carrying the Hfe∆CD-Myc transgene. Hfe∆CD-Myc lacks the C-terminal cytoplasmic domain of HFE but is able to induce hepcidin expression and to interact with TfR1 in livers.18,24 We found that Hfe∆CD-Myc also interacted with endogenous ALK3 in mouse livers (Figure 2F). These results demonstrate that HFE interacts with the BMP type I receptors ALK3 and ALK6.
ACTRIIA is the predominant BMP type II receptor in human livers.7 To investigate whether HFE interacts with ACTRIIA, we transfected Hep3B cells with ACTRIIA-Myc in combination with HFE-HA or ALK3-HA, and precipitated the lysates with HA antibody. There was no ACTRIIA-Myc band in the western blot in HFE-HA precipitates. As a positive control, there was an ACTRIIA band pulled down by ALK3-HA (Figure 2G; compare lanes 3 and 2). These results suggest that HFE does not interact with the BMP type II receptor ACTRIIA.
HFE and ALK3 synergistically enhance hepcidin expression
To examine whether HFE facilitates ALK3 signaling, we transfected Hep3B cells with ALK3 alone, HFE alone, and ALK3 in combination with HFE. ALK3 or HFE significantly increased hepcidin expression. Co-transfection of HFE and ALK3 further increased hepcidin expression, and this increase was more than the addition of the increases in hepcidin levels by ALK3 alone and by HFE alone (Figure 2H). These results suggest that HFE and ALK3 have synergistic effects on inducing hepcidin expression.
HFE increases ALK3 protein expression
We observed from our immunoprecipitation experiments that ALK3-HA expression was increased when HFE-Myc was coexpressed in COS7 (Figure 2B) and Hep3B (Figure 2D) cells. These results suggested that the HFE-ALK3 interaction might stabilize the ALK3 protein.
To further examine whether HFE increases ALK3 protein expression, we transfected Hep3B cells with a constant amount of ALK3-HA and increasing amounts of HFE-Myc. As determined by western blotting using the anti-HA antibody, ALK3 protein levels in the whole-cell lysates were elevated as HFE expression increased (Figure 3A), whereas ALK3 mRNA levels were not altered by HFE overexpression (Figure 3B). Similar results were observed in HepG2 cells when ALK3-HA and HFE-Myc plasmids were used (supplemental Figure 2A), and in Hep3B cells when ALK3-Flag and HFE-HA plasmids were used (supplemental Figure 2B). These results suggest that HFE stabilizes ALK3 protein in cultured cells.
HFE increases ALK3 protein expression in vitro and in vivo. (A) HFE increases ALK3 protein expression in Hep3B cells. Cells were transfected with and without a constant amount of ALK3-HA plasmid in the presence of increasing amounts of HFE-Myc. Forty-eight hours after transfection, cells were harvested for western blotting for ALK3 and HFE protein levels. (B) HFE does not alter ALK3 mRNA expression. Hep3B cells were transfected with and without ALK3-HA in the presence or absence of HFE-Myc. Forty-eight hours after transfection, cells were harvested for real-time PCR analysis for ALK3 mRNA levels. (C) HFE increases endogenous ALK3 protein expression in Hep3B cells. Cells were transfected with or without ALK3-HA in the presence or absence of HFE-HA plasmid. Forty-eight hours after transfection, cells were harvested for western blotting for endogenous ALK3 and transfected ALK3-HA protein using an ALK3 antibody from Millipore (left panel). The bands were quantified by densitometry (right panel). (D) ALK3 protein expression in WT and Hfe knockout (KO) mice. Liver lysates from WT and Hfe KO mice at 8 to 10 weeks of age were used for western blotting for ALK3 protein. Livers from Alk3fl/fl; Alb-Cre and the littermates Alk3fl/fl were included to determine the specific ALK3 protein bands (n = 3). The bands were quantified by densitometry (bottom panel). (E) Livers from WT and Hfe KO mice were analyzed for ALK3 mRNA expression. Rpl19 or RPL19 are the internal controls for real-time PCR. *P < .05, **P < .01. Data are represented as mean ± SD (n = 3 for panels B-D; n = 4 for panel E). All experiments were repeated 2 to 3 times.
HFE increases ALK3 protein expression in vitro and in vivo. (A) HFE increases ALK3 protein expression in Hep3B cells. Cells were transfected with and without a constant amount of ALK3-HA plasmid in the presence of increasing amounts of HFE-Myc. Forty-eight hours after transfection, cells were harvested for western blotting for ALK3 and HFE protein levels. (B) HFE does not alter ALK3 mRNA expression. Hep3B cells were transfected with and without ALK3-HA in the presence or absence of HFE-Myc. Forty-eight hours after transfection, cells were harvested for real-time PCR analysis for ALK3 mRNA levels. (C) HFE increases endogenous ALK3 protein expression in Hep3B cells. Cells were transfected with or without ALK3-HA in the presence or absence of HFE-HA plasmid. Forty-eight hours after transfection, cells were harvested for western blotting for endogenous ALK3 and transfected ALK3-HA protein using an ALK3 antibody from Millipore (left panel). The bands were quantified by densitometry (right panel). (D) ALK3 protein expression in WT and Hfe knockout (KO) mice. Liver lysates from WT and Hfe KO mice at 8 to 10 weeks of age were used for western blotting for ALK3 protein. Livers from Alk3fl/fl; Alb-Cre and the littermates Alk3fl/fl were included to determine the specific ALK3 protein bands (n = 3). The bands were quantified by densitometry (bottom panel). (E) Livers from WT and Hfe KO mice were analyzed for ALK3 mRNA expression. Rpl19 or RPL19 are the internal controls for real-time PCR. *P < .05, **P < .01. Data are represented as mean ± SD (n = 3 for panels B-D; n = 4 for panel E). All experiments were repeated 2 to 3 times.
To determine whether HFE increases endogenous ALK3 proteins, we transfected Hep3B cells with and without HFE-HA, and the whole-cell lysates were subjected to western blotting using an anti-ALK3 antibody to detect endogenous ALK3 protein. Cells transfected with ALK3-HA were used as a reference for the ALK3 protein size. As is shown in Figure 3C, HFE overexpression increased endogenous ALK3 protein expression.
To examine whether HFE regulates ALK3 protein expression in vivo, we used Hfe knockout mice. Consistent with the previous observation,26 hepatic phospho-Smad1/5/8 relative to Bmp6 mRNA was significantly reduced in Hfe knockout compared with WT mice (supplemental Figure 3), indicating defective BMP signaling in Hfe knockout mice.26 Hepatic ALK3 protein levels were significantly reduced in Hfe knockout mice compared with WT mice as is shown by western blotting (Figure 3D). As a control for antibody specificity, we found that the ALK3 bands were much reduced in liver samples from mice with a liver-specific deletion of Alk3 (Alk3fl/fl; Alb-Cre) compared with the control littermates (Alk3fl/fl, Figure 3D). Interestingly, hepatic Alk3 mRNA levels were similar between WT and Hfe knockout mice (Figure 3E). These results suggest that HFE may increase ALK3 protein expression in vivo.
HFE stabilizes the ALK3 protein by inhibiting its ubiquitination proteasomal degradation
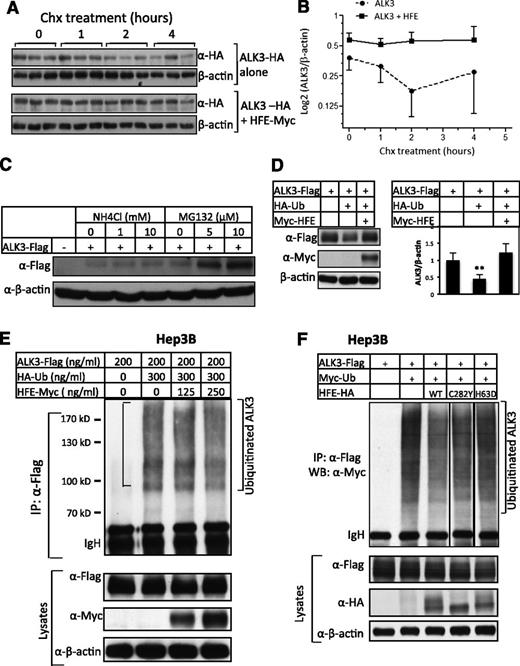
To explore the mechanism by which HFE stabilizes ALK3 protein, we first studied whether HFE inhibits ALK3 protein degradation. Hep3B cells were transfected with ALK3-HA in the presence or absence of HFE-Myc. Cells were then treated with Chx for 0, 1, 2, and 4 hours before being analyzed for ALK3 protein levels. As is shown in Figure 4A-B, ALK3 protein levels dramatically declined from 0 to 2 hours after Chx treatment in the absence of HFE, whereas ALK3 proteins remained unchanged in the presence of HFE. These results suggest that HFE inhibits ALK3 degradation.
HFE stabilizes ALK3 protein by inhibiting its ubiquitination and proteasomal degradation. (A-B) HFE inhibits ALK3 degradation. Hep3B cells were transfected with ALK3-HA in the presence or absence of HFE-Myc. Forty-six hours later, cells were incubated with Chx for 0, 1, 2, and 4 hours. Cells were then harvested and subjected to western blot analysis for ALK3 protein levels using anti-HA antibody. A representative western blot is shown in (A). Densitometric analysis is shown in (B). (C) ALK3 degrades primarily through the proteasomal pathway. Hep3B cells were transfected with empty vector or ALK3-Flag (200 ng/mL). Forty-six hours after transfection, cells were treated with increasing doses of NH4Cl (0, 1, and 10 mM) or MG132 (0, 5, and 10 μM) for 5 hours. Whole-cell lysates were prepared for western blot analysis with anti-Flag antibody for ALK3 protein expression. (D) HFE attenuates ubiquitin-mediated ALK3 degradation. Hep3B cells were transfected with ALK3-Flag alone, or ALK3-Flag in combination with HA-ubiquitin in the absence or presence of HFE-Myc. Forty-eight hours after transfection, cells were harvested for western blotting for ALK3 and HFE protein levels. Representative western blot is shown in the left panel, and quantification of 3 separate experiments is shown in the right panel. (E) HFE inhibits ALK3 ubiquitination. Hep3B cells were transfected with ALK3-Flag alone, or ALK3-Flag in combination with HA-ubiquitin in the presence of increasing amounts of HFE-Myc. Forty-six hours after transfection, cells were treated with MG132 (5 μM) for 5 hours. Whole-cell lysates were precipitated with anti-Flag antibody and analyzed by western blotting for ubiquitinated ALK3 using anti-HA antibody. Whole-cell lysates were also used for western blot analysis for AKL3 and HFE expression. (F) HFE-H63D does not inhibit ALK3 ubiquitination. Hep3B cells were transfected with ALK3-Flag alone, or ALK3-Flag in combination with Myc-ubiquitin in the presence of HFE-HA (WT), HFE-HA (C282Y), or HFE-HA (H63D). Forty-six hours after transfection, cells were treated with MG132 (5 μM) for 5 hours. Whole-cell lysates were precipitated with anti-Flag antibody and analyzed by western blotting for ubiquitinated ALK3 using anti-Myc antibody. Whole-cell lysates were also used for western blot analysis for AKL3 and HFE expression. β-actin is the loading control. *P < .05, **P < .01. Data are represented as mean ± SD (n = 3 for panels A-B). All the experiments were repeated 2 to 3 times.
HFE stabilizes ALK3 protein by inhibiting its ubiquitination and proteasomal degradation. (A-B) HFE inhibits ALK3 degradation. Hep3B cells were transfected with ALK3-HA in the presence or absence of HFE-Myc. Forty-six hours later, cells were incubated with Chx for 0, 1, 2, and 4 hours. Cells were then harvested and subjected to western blot analysis for ALK3 protein levels using anti-HA antibody. A representative western blot is shown in (A). Densitometric analysis is shown in (B). (C) ALK3 degrades primarily through the proteasomal pathway. Hep3B cells were transfected with empty vector or ALK3-Flag (200 ng/mL). Forty-six hours after transfection, cells were treated with increasing doses of NH4Cl (0, 1, and 10 mM) or MG132 (0, 5, and 10 μM) for 5 hours. Whole-cell lysates were prepared for western blot analysis with anti-Flag antibody for ALK3 protein expression. (D) HFE attenuates ubiquitin-mediated ALK3 degradation. Hep3B cells were transfected with ALK3-Flag alone, or ALK3-Flag in combination with HA-ubiquitin in the absence or presence of HFE-Myc. Forty-eight hours after transfection, cells were harvested for western blotting for ALK3 and HFE protein levels. Representative western blot is shown in the left panel, and quantification of 3 separate experiments is shown in the right panel. (E) HFE inhibits ALK3 ubiquitination. Hep3B cells were transfected with ALK3-Flag alone, or ALK3-Flag in combination with HA-ubiquitin in the presence of increasing amounts of HFE-Myc. Forty-six hours after transfection, cells were treated with MG132 (5 μM) for 5 hours. Whole-cell lysates were precipitated with anti-Flag antibody and analyzed by western blotting for ubiquitinated ALK3 using anti-HA antibody. Whole-cell lysates were also used for western blot analysis for AKL3 and HFE expression. (F) HFE-H63D does not inhibit ALK3 ubiquitination. Hep3B cells were transfected with ALK3-Flag alone, or ALK3-Flag in combination with Myc-ubiquitin in the presence of HFE-HA (WT), HFE-HA (C282Y), or HFE-HA (H63D). Forty-six hours after transfection, cells were treated with MG132 (5 μM) for 5 hours. Whole-cell lysates were precipitated with anti-Flag antibody and analyzed by western blotting for ubiquitinated ALK3 using anti-Myc antibody. Whole-cell lysates were also used for western blot analysis for AKL3 and HFE expression. β-actin is the loading control. *P < .05, **P < .01. Data are represented as mean ± SD (n = 3 for panels A-B). All the experiments were repeated 2 to 3 times.
We then explored which pathway is involved in ALK3 degradation. We incubated Hep3B cells transfected with ALK3-Flag with increasing doses of the proteasome inhibitor MG132 or the lysosome inhibitor NH4Cl. As is shown in Figure 4C, MG132 increased ALK3 protein expression dose-dependently, whereas NH4Cl at concentrations of up to10 mM had very small effects, suggesting that ALK3 is primarily degraded through proteasomes.
We also examined whether ubiquitination plays a role in ALK3 degradation. ALK3 protein expression was inhibited in the presence of HA-ubiquitin compared with the ALK3 expression in the absence of HA-ubiquitin (Figure 4D). Overexpression of HFE attenuated the inhibition of ALK3 protein expression by ubiquitin (Figure 4D). To directly examine the effect of HFE on polyubiquitinated ALK3 levels, we transfected Hep3B cells with a constant amount of ALK3-Flag in the presence of a constant amount of HA-ubiquitin and increasing amounts of HFE-Myc, and then treated the cells with MG132. Polyubiquitinated ALK3 was readily detected by western blotting using anti-HA antibody in the anti-Flag antibody precipitates (Figure 4E, lane 2). HFE cotransfection inhibited ALK3 ubiquitination (Figure 4E; compare lanes 3 and 4 with lane 2). Therefore, our results suggest that ALK3 degrades through the ubiquitination-proteasomal pathway, and this process is inhibited by HFE.
HFE increases ALK3 expression on the cell surface
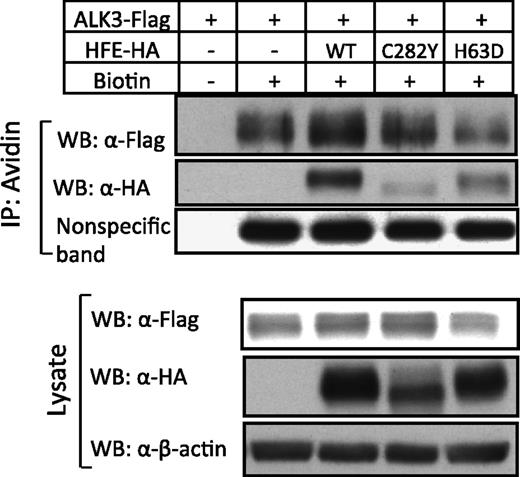
To examine whether the increased levels of ALK3 protein by HFE is transported to the cell surface, we performed cell-surface biotinylation on whole cells transfected with ALK3-Flag alone or ALK3-Flag in combination with HFE-HA. As is shown in Figure 5, overexpression of HFE increased ALK3 protein expression on the cell surface (compare lanes 3 and 2, top panel), as well as in the whole-cell lysates (compare lanes 2 and 1, bottom panel). As a loading control, the expression of a nonspecific membrane protein was not altered by HFE in biotinylated surface proteins. HFE surface expression was detected in cells transfected with HFE-Myc (lane 3, top panel). As a negative control, no ALK3 in streptavidin precipitates was detected when the cells were not biotinylated (lane 1, top panel). These results suggest that HFE is able to increase membrane accumulation of ALK3 in Hep3B cells.
HFE, but not HFE-C282Y and HFE-H63D, increases ALK3 cell-surface expression. Hep3B cells were transfected with ALK3-Flag in the presence or absence of HFE-HA (WT), HFE-HA (C282Y), and HFE-HA (H63D). Forty-eight hours after transfection, whole cells were incubated with and without Sulfo-NHS-LC-LC-Biotin, and the cell lysates were precipitated with Streptavidin Agarose. The whole-cell lysates and avidin precipitates were subjected to western blot analysis with antibodies as indicated. A nonspecific membrane protein was used as loading control. The experiment was repeated 3 times.
HFE, but not HFE-C282Y and HFE-H63D, increases ALK3 cell-surface expression. Hep3B cells were transfected with ALK3-Flag in the presence or absence of HFE-HA (WT), HFE-HA (C282Y), and HFE-HA (H63D). Forty-eight hours after transfection, whole cells were incubated with and without Sulfo-NHS-LC-LC-Biotin, and the cell lysates were precipitated with Streptavidin Agarose. The whole-cell lysates and avidin precipitates were subjected to western blot analysis with antibodies as indicated. A nonspecific membrane protein was used as loading control. The experiment was repeated 3 times.
HFE-C282Y and HFE-H63D interact with ALK3 but fail to increase ALK3 surface expression
As is shown in Figure 2D, like WT HFE, HFE-C282Y or HFE-H63D also interacted with ALK3. Interestingly, although WT HFE and HFE-C282Y inhibited ALK3 ubiquitination, HFE-H63D failed to reduce the ubiquitinated ALK3 levels (Figure 4F). These results suggest that, in addition to the interaction between HFE and ALK3, some other features in the HFE protein are required for HFE to inhibit ALK3 ubiquitination.
Consistent with their ability to inhibit ALK3 ubiquitination, wild-type HFE and HFE-C282Y increased ALK3 protein expression in whole lysates of Hep3B cells, but HFE-H63D did not (Figure 2D and Figure 5, lower panel). Interestingly, despite the increased ALK3 expression in whole lysates, ALK3 surface expression was not increased by HFE-C282Y compared with cells transfected with ALK3 alone (Figure 5; compare lanes 4 and 2, top panel). In addition, HFE-H63D did not increase ALK3 surface expression either (Figure 5; compare lanes 5 and 2, top panel). As expected, HFE-H63D was detected on the cell surface, but HFE-C282Y was barely detectable (Figure 5, top panel).
To corroborate the results from cell-surface labeling, we performed confocal immunofluorescence analysis on Hep3B cells transfected with ALK3-Flag alone or ALK3-Flag in combination with HFE-HA WT, HFE-HA C282Y, or HFE-HA H63D (supplemental Figure 4). Transfection of HFE-WT dramatically increased ALK3 expression on the cell surface compared with cells transfected with ALK3 alone. Transfection of HFE-C282Y or HFE-H63D did not increase ALK3 expression on the cell surface.
Discussion
It has been well-documented that HFE, TFR2, HJV, and BMP6 are critical activators of hepcidin expression in the liver. Although our previous studies showed that HJV functions as a BMP coreceptor to stimulate hepcidin expression, the mechanisms by which HFE and TFR2 regulate hepcidin expression remain unsolved. Interestingly, defective BMP/Smad signaling has been observed in Hfe and Tfr2 knockout mice16,19,26,27 and in HFE-HH patients.28,29 These data implicate the BMP pathway in HFE- and TFR2-regulated hepcidin expression. In the present study, we provide direct biochemical evidence that HFE regulates hepcidin expression through the BMP pathway. We first observed 1.3- to 2.2-fold increases in hepcidin expression by HFE overexpression in Hep3B cells in our culture system. These changes are comparable with the 1.7- to 1.8-fold increases in hepcidin expression by virus-mediated liver-specific overexpression of HFE17 or the 1.9-fold increase in hepcidin expression by transgenic overexpression of HFE in mice.18 We then demonstrated that HFE overexpression increased Smad1/5/8 phosphorylation in Hep3B cells, whereas disease-causing mutants HFE-C282Y and HFE-H63D failed to activate Smad1/5/8 and had no or little effect on hepcidin expression. Furthermore, HFE-induced hepcidin expression was abrogated by treatment of the BMP inhibitor LDN-193189. In addition, we revealed that HFE interacted with ALK3 in cells and in livers, and that HFE and ALK3 synergistically increased hepcidin expression. All of these results suggest that HFE can act through ALK3 to influence the BMP pathway and hepcidin expression.
Consistent with previous studies,34,35 our results demonstrated that ALK3 undergoes the ubiquitination-proteasome degradation pathway. HFE overexpression inhibited ALK3 ubiquitination and degradation. The mechanism by which HFE inhibits ALK3 ubiquitination remains unknown. A previous study showed that Smad ubiquitin regulatory factor (Smurf) 1 binds to BMP type I receptors through the inhibitory Smads (I-Smad), Smad6 and Smad7, and induces ubiquitination of these receptors.34 Whether HFE inhibits ALK3 ubiquitination by interfering with the formation of the Smurf1-I-Smads-ALK3 complex is an intriguing question to be addressed.
Consistent with the increased ALK3 protein expression in whole-cell lysates, membrane accumulation of ALK3 was also enhanced by HFE. This at least partially explains the role of HFE in regulating BMP signaling and hepcidin expression. However, whether and how HFE interacts with ALK3 to regulate BMP signaling once they are present on the cell surface remains to be investigated. Interestingly, Hfe knockout mice exhibited decreased ALK3 protein expression in the liver compared with WT mice, whereas Alk3 mRNA levels are similar between the 2 genotypes. This suggests that HFE may activate hepcidin expression by increasing ALK3 protein expression in vivo.
HFE-C282Y and HFE-H63D interacted with ALK3 but did not induce hepcidin expression in Hep3B cells, implying that in disease states, HFE and ALK3 interact but the subsequent biochemical processes leading to activation of the BMP pathway are impaired. Unlike WT HFE, HFE-H63D was not able to inhibit ALK3 ubiquitination and thus failed to increase ALK3 protein expression in whole-cell lysates and on the cell surface. Surprisingly, although HFE-C282Y inhibited ALK3 ubiquitination and increased ALK3 protein expression in whole-cell lysates, this mutant did not increase ALK3 surface expression. Previous studies revealed that the C282Y mutant does not associate with β2-microglobulin, so that it does not traffic to the cell surface.31-33 Consistent with this finding, we did not observe significant HFE282Y cell-surface expression. Therefore, it is possible that HFE-C282Y may sequester ALK3 inside cells, thus preventing ALK3 from trafficking to the cell surface. Our results also suggest that normal HFE trafficking is required for ALK3 localization.
We observed a similar interaction of HFE and ALK6. However, ALK6 is not expressed in mouse13 or human liver,7 so the interaction of HFE and ALK6 could be important to other organs but is likely not relevant to hepatic physiology.
Tfr2 knockout mice failed to increase Smad1/5/8 phosphorylation and hepcidin expression, despite iron overload and increased BMP6 expression.19 These results suggest that TFR2 is also required for normal response of BMP signaling and hepcidin expression to iron status. However, how TFR2 activates hepcidin expression remains elusive. In the present study, we found that TFR2 exhibited a weak interaction with ALK3 (supplemental Figure 5), but transfection of TFR2 failed to stimulate Smad1/5/8 phosphorylation and hepcidin expression (supplemental Figure 5). The mechanisms by which TFR2 regulates hepcidin expression are still under investigation in our laboratory.
HFE and TFR2 also interact with HJV.23 Therefore, it is possible that HFE, TFR2, HJV, and ALK3 form a complex, and within this complex HFE, TFR2, and HJV may receive signals such as transferrin-bound iron levels and convey them to ALK3, thereby regulating the downstream Smad activities and hepcidin expression.
Taken together, our present study has shown that HFE induces hepcidin expression via the BMP pathway. HFE interacts with ALK3, increasing ALK3 protein expression both in vitro and in vivo. HFE stabilizes ALK3 protein by inhibiting the ubiquitination proteasomal degradation of ALK3. HFE increases ALK3 cell-surface expression. Disease-causing HFE mutations may impair HFE ability in inhibiting ALK3 degradation or in tethering ALK3 to the cell surface.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Nancy Andrews for the Hfe−/− mice; Drs Herbert Lin and Jodie Babitt for their careful and critical reading of the manuscript; Dr Hui Zhao and Dr Michael S.Y. Huen for the HA-ubiquitin and Myc-ubiquitin plasmids, respectively; and Dr John Fleming for HFE-HA (WT) and HFE-HA (C282Y) constructs. The HFE-Myc and TFR2-myc/his plasmids were generated by Dr Paul Schmidt in the laboratory of Dr Nancy C. Andrews and provided by Dr Diedra M. Wrighting. Dr Kohei Miyazono provided the ALK3-HA plasmid.
This study was supported by the startup fund offered by The Chinese University of Hong Kong (CUHK), an RGC/GRF grant (CUHK477311), an RGC-NSFC joint grant (N_CUHK432/12 and 81261160507), CUHK direct grants (2041603 and 2041747), and Shenzhen Science and Technology Research and Development Funding (JC201105201069A) (Y.X.); grants from The National Natural Science Foundation of China (31030039 and 31225013) (F.W.); and CUHK postgraduate scholarships (X.-G.W., W.L., Y.Z.).
Authorship
Contribution: Y.X. designed the research, analyzed the data and wrote the paper; X.W. designed the research, performed the research, analyzed data, and wrote the paper; Y.W. and W.C. performed the research; and C.M., P.J.S., P.B.Y., F.W., and Q.W. provided key reagents and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Yin Xia, School of Biomedical Sciences, The Chinese University of Hong Kong, Shatin, N.T., Hong Kong; e-mail: xia.yin@cuhk.edu.hk.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal