Key Points
VKORC1:p.Arg98Trp disrupts a di-arginine ER retention motif, resulting in mislocalization and degradation of the mutant VKORC1 protein.
A second low-efficiency di-lysine ER localization and retention motif contributes to the partially deficient phenotype of VKCFD2 patients.
Abstract
Vitamin K 2,3-epoxide reductase complex subunit 1 (VKORC1) is an enzyme localized to the endoplasmic reticulum (ER) membrane. VKORC1 catalyzes the reduction of vitamin K 2,3-epoxide to vitamin K and to vitamin K hydroquinone, the latter required by the enzyme γ-carboxylase for γ-carboxylation of all vitamin K–dependent (VKD) proteins. Until now, only 1 human VKORC1 mutation, p.Arg98Trp, is known to cause combined deficiency of VKD clotting factors type 2 (VKCFD2), a disease phenotype reported in 3 unrelated families. VKCFD2 patients suffer from spontaneous bleeding episodes because of decreased levels of γ-carboxylated VKD clotting factors. Daily supraphysiological vitamin K supplementation restores clotting for VKCFD2 patients and results in high serum levels of vitamin K 2,3-epoxide, suggesting that supplemented vitamin K is reduced in vivo. Although the p.Arg98Trp mutation results in reduced vitamin K 2,3-epoxide reductase activity, the molecular mechanism underlying this pathophysiology is unknown. Using a combination of in silico analysis and confocal microscopy, we demonstrate for the first time that VKORC1:p.Arg98Trp disrupts a di-arginine ER retention motif resulting in 20% ER colocalization only. As a consequence, VKORC1 exits the ER membrane by cellular quality control systems and results in the observed VKCFD2 phenotype.
Introduction
Vitamin K 2,3-epoxide reductase complex subunit 1 (VKORC1) is an endoplasmic reticulum (ER) transmembrane protein that is the rate-limiting enzyme of the vitamin K cycle.1,2 VKORC1 catalyzes the reduction of vitamin K 2,3-epoxide (K > O) to vitamin K quinone (K) and, additionally, to vitamin K hydroquinone (KH2). The KH2 generated is the substrate for the second essential enzyme of the vitamin K cycle, γ-glutamyl carboxylase, which posttranslationally modifies vitamin K–dependent (VKD) proteins. This modification enables binding of calcium ions essential for physiological function of VKD proteins. During γ-carboxylation, KH2 is oxidized to vitamin K 2,3-epoxide to complete the vitamin K cycle.3,4
Defects in the vitamin K cycle can cause combined deficiency of VKD clotting factors (VKCFD), resulting in reduced activity levels for VKCFD FII, FVII, FIX, and FX, as well as for other VKD proteins. This deficiency can be caused either by various mutations in γ-glutamyl carboxylase leading to VKCFD type 1 or by a single VKORC1 missense mutation that results in VKCFD type 2 (VKCFD2)1,5 The VKCFD2 phenotype was first presented by Pauli et al,6 followed by a more detailed description of 7 patients from 2 independent families by Oldenburg et al.7 The latter 2 families enabled a homozygosity mapping study locating the affected gene for this phenotype to chromosome 16.8 Finally, in 2004, the VKORC1 gene was identified, and all affected individuals from both VKCFD2 families were found to carry the same homozygous VKORC1:p.Arg98Trp mutation.1,2 Currently, p.Arg98Trp is the only VKORC1 mutation known to cause VKCFD2.1,5 Afflicted patients exhibit VKD coagulation factor activities in the range of 20% to 60% compared with patients with wild-type (wt) VKORC1.6,7 This phenotype can be successfully treated by daily vitamin K supplementation (1-10 mg) that leads to functional VKD coagulation factor activities in the normal range of 70-130 U/dL.5-7 Vitamin K supplementation for VKCFD2 patients is associated with a high serum concentration of vitamin K 2,3-epoxide (19.5-66.2 ng/mL).7 Although in healthy individuals K > O is undetectable in serum even after administration of 10 mg vitamin K,9 the mechanism by which vitamin K supplementation rescues VKD coagulation factor levels is not understood. In addition, in vitro investigation by the dithiothreitol-driven vitamin K 2,3-epoxide reductase assay reveals significantly reduced vitamin K 2,3-epoxide reductase activity of only 9%, for the VKORC1:p.Arg98Trp variant compared with that of wt.1
Using a combination of in silico analysis and confocal microscopy, we demonstrate for the first time that this mutation abrogates an ER retention motif responsible for the correct intracellular localization of VKORC1. Our results also suggest that 20% of the mutant protein VKORC1:p.Arg98Trp is retained in the ER, possibly explaining the ability of daily supplemented vitamin K to rescue the VKCFD2 phenotype in vivo.
Methods
Motif search and primary structural analysis of the region around p.Arg98Trp
We had previously published a homology model of human VKORC1 based on the bacterial homolog crystal structure (PMDB database: http://bl209.frm.uniroma1.it/PMDB/; accession ID: PM0079536).10,11 We performed membrane-embedded simulation for this model using a macro plug-in embedded in YASARA version 13.8.6.12 For equilibration, a phosphatidylethanolamine membrane was used, and the membrane size was determined in YASARA by first determining the transmembrane regions and then fine-tuning manually the membrane position with respect to it. A compression simulation shrank the simulation cells fusing the membrane edges with the periodic boundaries, which resulted in a final membrane size of 67X66 Å. The compressed protein was then embedded in the phosphatidylethanolamine membrane, and the lipids that bumped into the protein were deleted. The protein was then slowly expanded during a simulation to fill the pore. The simulation cell was filled with water, carefully avoiding placing water molecules between the lipids, and the whole system was energy minimized to remove any conformational stress from bumps. Postembedding, a 250-ps equilibration simulation run was performed during which the membrane stabilizes and adjusts to the protein. Finally, we used this membrane-embedded model to inspect the predicted protein structure near the Arg98 residue. We also conducted a linear motif search for the VKORC1 protein sequence on the eukaryotic linear motif (ELM) database (http://elm.eu.org/).13 Search criteria were limited by species to Homo sapiens. Because the VKORC1 homology model provided us important structural insights, we considered the complete set of results from the motif search including excluded ELMs with poor structural filter scores. A multiple alignment of VKORC1 homologs was downloaded from the Homologene database (http://www.ncbi.nlm.nih.gov/homologene) to assess conservation status for the identified retention motifs and surrounding sequences.14
Mammalian expression vectors
Human VKORC1 (hVKORC1) complementary DNA was cloned into the pEGFP-N3 vector (Clontech, Saint-Germain-en-Laye, France) by a restriction-free polymerase chain reaction protocol using Phusion DNA polymerase (Finnzymes, Vantaa, Finnland).15 In this vector, enhanced green fluorescent protein (EGFP) is C-terminal tagged to VKORC1 for all variants investigated in this study. Site-directed mutagenesis was performed using PFU-Turbo-polymerase (Agilent Technologies, Böblingen, Germany). Mutant variants were generated in the pEGFP-N3 wt VKORC1 containing vector for the p.Arg98Trp mutation as well as for the critical residues suspected to influence the identified ER retention motifs. VKORC1 variant constructs for expression and investigation included the following: (1) for arginine-dependent motifs: Arg33Ala + Arg35Ala, Arg98Ala + Arg100Ala, 98_100delRTR, p.Arg98Trp; (2) for lysine-dependent motifs: Lys159Ala + Lys161Ala; and (3) for combination variants: Arg98Ala + Arg100Ala + Lys159Ala + Lys161Ala, 98_100delRTR + Lys159Ala + Lys161Ala, p.Arg98Trp + Lys159Ala + Lys161Ala. See supplemental Table 1 (see the Blood Web site) for mutagenesis primers.
Immunofluorescent confocal microscopy
HEK293T cells were plated on cover glasses and transfected with Fugene HD transfection reagent according to the manufacturer’s protocol (Promega, Madison, WI). Twenty-four hours posttransfection, cells were washed with phosphate buffered saline (PBS), fixed with 4% paraformaldehyde for 10 minutes at room temperature (RT), and afterward incubated with blocking solution (89.9% PBS, 10% fetal bovine serum, 0.1% Triton) for 30 minutes at RT.
For ER staining, cells were stained for 2 hours at RT with an anti–protein disulfide isomerase (PDI) antibody (Invitrogen, Darmstadt, Germany), diluted in antibody solvent solution (98.9% PBS, 1% fetal bovine serum, 0.1% Triton-X100). As primary antibody for staining the Golgi apparatus, anti-Golgin-97 was used with a concentration of 100 µg/mL (Invitrogen). After incubation with one of the primary antibodies, cover glasses were washed twice with PBS. Secondary antibody was an anti-primary AlexaFluor 594 conjugate at 1:1000 dilution for 1 hour (Invitrogen, Darmstadt, Germany).
The plasma membrane was stained using CellMask Orange plasma membrane stain (Invitrogen) before fixation as recommended by the manufacturer’s protocol by adding 5 µg/mL CellMask into the medium. After 5 minutes incubation at 37°C with CellMask, cells were washed with PBS and fixed with 4% paraformaldehyde.
The nucleus was counterstained in all experiments for 3 minutes with ToPro3 iodide (Invitrogen), diluted 1:10 000 in blocking solution. All preparations were washed 3 times with PBS after immunostaining. Finally, cover glasses were embedded in VectaShield mounting medium (Vector Laboratories, Burlingame, CA) and covered with a coverslip. Observation and imaging were performed with an Olympus Fluoview FV 1000 confocal laser scanning microscope using a ×40 water immersion Fluor lens and Olympus Fluoroview 1.4 software. Emitted fluorescence was detected by an Olympus camera. Excitation maxima for PDI and Golgi staining were 594 nm; for plasma membrane, 549 nm; for GFP-tagged VKORC1, 488 nm; and for the nucleus, 642 nm. The photomultiplier tube voltage for GFP had to be increased from 400 V to 800 V for all hVKORC1 variants compared with wt VKORC1 and the Arg33Ala + Arg35Ala variant.
Image analysis
Colocalization analysis was performed using plug-ins embedded in the visualization and analysis software ImageJ 1.43m.16 Analysis was performed on a similar-sized symmetrical region of interest (ROI) selected for each dye. Background levels were subtracted from each ROI before calculating the degree of colocalization (to a range of 1 standard deviation [SD]). Each colored image was split into respective red, green, and blue channels. The comparative degree of colocalization for the wt and mutant variants was calculated as mean Pearson’s and Mander’s R coefficients on the red and green channels using the embedded colocalization analysis plug-in at default settings.17 The colocalization highlighter plug-in was used with default setting (50% threshold values for both channels) to further visualize the colocalized pixels rendered as white. Because Pearson’s and Mander’s R coefficients showed good correlation (Mander’s R was almost consistently higher than Pearson’s correlation coefficient), Pearson’s correlation coefficient was used for comparing relative degree of colocalization between the mutant variants and wt.18 A total of n = 18-50 ROIs were evaluated for each pairwise comparison. The lower number of ROIs investigated corresponds to the deletion variants in which very few cells were actually observed to express the variant protein.
The number of cells expressing the EGFP-tagged hVKORC1 variant per view was calculated as the ratio of the total number of cells observed in a view (×40) to the number of cells emitting fluorescent signal from the expressed protein. Five images of 3 different transfections and stainings were counted for each hVKORC1 variant investigated. The ratio was normalized per 500 cells. Means were compared using the parametric Student t test; a P value <.05 was considered as significant.
Results
Homology model inspection and sequence motif search
Motif search for the hVKORC1 amino acid sequence in the ELM database uncovered 2 putative ER retention motifs (supplemental Figure 1). One was a di-lysine motif (KAKRH; ELM motif ID:TRG_ER_diLys_1) at the C terminus of wt hVKORC1 (Lys159_His163), and the other was a di-arginine motif LRTR (Leu97_Arg100) or RTRW (Arg98_Trp101) or RTR (Arg98_Arg100; ELM motif ID: TRG_ER_diArg_1) predicted to be in the first cytoplasmic loop between the second and third transmembrane α-helix. The di-lysine and di-arginine motifs had high probability scores (<0.0001 and 0.00524, respectively), indicating that both sequence motifs are likely localization and retention signals.
The di-arginine motif includes the Arg98 residue for which the missense mutation to tryptophan causes VKCFD2. In addition, inspection of the homology model (Figure 1) around the Arg98 residue shows that (1) this region is at the cytoplasmic interface of the ER membrane connecting the second and third transmembrane helices and that (2) the Arg98 residue is ∼15 Å (12-17 Å) distant from the membrane surface and orientated toward the cytoplasm.19 This predicted spatial positioning of the motif with respect to the cytoplasm/membrane surface suggests that the di-arginine motif could be a potential ER retention signal. Furthermore, 3 similar di-arginine ER retention motifs (LRTR, RTRW, and RTR) corresponding to linear sequences of hVKORC1 are found in database entries. Among the 27 instances of RXR motifs found in the ELM database, 9 belong to the LRTR category (XRXR), 6 to the RTRW (RXRX) category, and 8 to the core RTR (RXR) category (supplemental Table 2). The di-lysine motif observed in hVKORC1 also fulfilled the typical characteristics observed in known di-lysine ER retention motifs.20 Accordingly, it is located at the C terminus, and there is a minimum spacing of 5 amino acids between any adjacent transmembrane domain and the motif. In the ELM search, a total of 12 di-lysine motifs are found for the KKXX and the KXKXX categories (VKORC1 belongs to the KXKXX category; supplemental Table 3). Three out of the 12 database entries for the di-lysine motif in the ELM database belong to the KXKXX category.
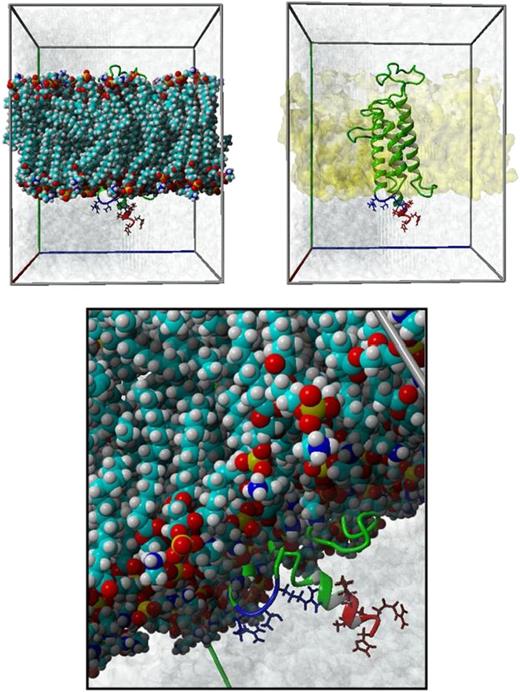
Localization of Arg98 in the membrane-embedded homology-based model of hVKORC1 protein. This figure illustrates the spatial positioning of the ER retention motifs with respect to the ER membrane in the membrane-embedded homology model of hVKORC1 protein. The upper 2 panels show the membrane-embedded hVKORC1 protein in a simulation cell. The ER membrane is represented with ball models on the right and by its molecular surface (yellow colored) on the left. The hVKORC1 protein is represented in ribbon form, whereas the residues, which are part of the ER retention motifs, are represented as stick models in red (di-lysine motif, Lys159_His163) and blue (di-arginine motif, Arg98_Arg100). The lower panel is a closer look at both motifs. The water molecules within the simulation box are represented by their molecular surface in blue.
Localization of Arg98 in the membrane-embedded homology-based model of hVKORC1 protein. This figure illustrates the spatial positioning of the ER retention motifs with respect to the ER membrane in the membrane-embedded homology model of hVKORC1 protein. The upper 2 panels show the membrane-embedded hVKORC1 protein in a simulation cell. The ER membrane is represented with ball models on the right and by its molecular surface (yellow colored) on the left. The hVKORC1 protein is represented in ribbon form, whereas the residues, which are part of the ER retention motifs, are represented as stick models in red (di-lysine motif, Lys159_His163) and blue (di-arginine motif, Arg98_Arg100). The lower panel is a closer look at both motifs. The water molecules within the simulation box are represented by their molecular surface in blue.
Manual inspection of the amino acid sequence of hVKORC1 suggests a second putative di-arginine motif at position Arg33_Arg35 (not observed during the ELM motif search). This second putative di-arginine motif would not fulfill the criteria for di-arginine ER retention motifs in our homology-based hVKORC1 model because of its location in the ER lumenal loop. However, in the literature there exists the possibility of a 3 transmembrane (3TM) topology-based model for hVKORC1.21 In this 3TM model, both di-arginine motifs are located in the cytoplasm and theoretically fulfill the characteristics of a di-arginine motif.
Colocalization analysis of the ER with wt and engineered di-arginine and di-lysine motif hVKORC1 variants in HEK293 cells
In the present study, we have mutated the arginines and lysine residues to neutral, small alanines for the 3 putative ER retention motifs (eg, Arg33Ala + Arg35Ala, Arg98Ala + Arg100Ala, Lys159Ala + Lys161Ala). We also generated 2 further variants for testing: a deletion construct removing the core di-arginine motif Arg98_Arg100 (98_100delRTR) and the human pathogenic variant p.Arg98Trp. The colocalization analysis showed variability in expression and ER retention. The wt hVKORC1 was almost exclusively localized to the ER as observed in the images and characterized by high colocalization coefficients (Figures 2 and 3). In addition, wt hVKORC1 showed the highest level of expression as indicated by the large number of hVKORC1-EGFP–expressing cells observed per analyzed image (126 of 500 cells, Figure 4). In comparison, general expression of the pEGFP vector without hVKORC1 complementary DNA was similar to the expression of wt hVKORC1 (109 of 500 cells, Figure 4). EGFP shows a ubiquitous expression in the nucleus and cytoplasm and no ER colocalization (supplemental Figure 3).
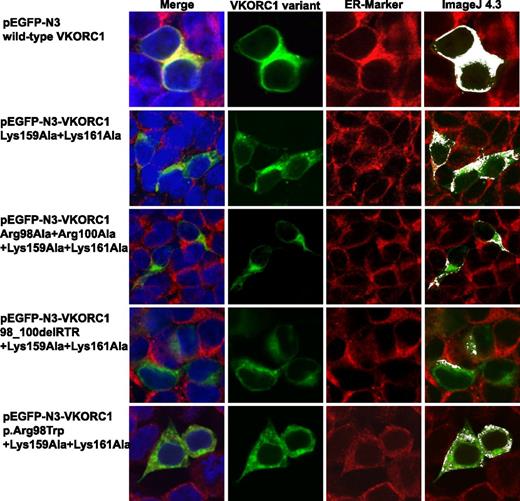
Colocalization images for wt hVKORC1 and mutated di-arginine motif variants in HEK293T cells. This figure shows representative colocalization images of single immunostained HEK293T cells expressing wt hVKORC1, Arg33Ala + Arg35Ala, Arg98Ala + Arg100Ala, the 98_100delRTR variant, and the human p.Arg98Trp mutant variant, respectively. Each panel is composed of 4 images; the first one is a merged picture of the green-colored EGPF-tagged hVKORC1 protein, the red-stained ER (primary antibody: anti-PDI; secondary antibody: AlexaFluor 594), and the blue-counterstained nucleus by ToPro3. Colocalization is represented with increasing intensity of yellow in the first image. The second image shows cells expressing the EGFP-tagged hVKORC1 protein in green only, followed by the third image in which the ER is red stained. The last column shows colocalized regions as white dots using the colocalization highlighter plug-in embedded in ImageJ 4.3. All images were rendered using ImageJ version 4.3. The images clearly show a high ER colocalization for wt hVKORC1 and the Arg33Ala + Arg35Ala variant. All other variants affecting the Arg98_Arg100 di-arginine motif show a drastic decrease in ER colocalization.
Colocalization images for wt hVKORC1 and mutated di-arginine motif variants in HEK293T cells. This figure shows representative colocalization images of single immunostained HEK293T cells expressing wt hVKORC1, Arg33Ala + Arg35Ala, Arg98Ala + Arg100Ala, the 98_100delRTR variant, and the human p.Arg98Trp mutant variant, respectively. Each panel is composed of 4 images; the first one is a merged picture of the green-colored EGPF-tagged hVKORC1 protein, the red-stained ER (primary antibody: anti-PDI; secondary antibody: AlexaFluor 594), and the blue-counterstained nucleus by ToPro3. Colocalization is represented with increasing intensity of yellow in the first image. The second image shows cells expressing the EGFP-tagged hVKORC1 protein in green only, followed by the third image in which the ER is red stained. The last column shows colocalized regions as white dots using the colocalization highlighter plug-in embedded in ImageJ 4.3. All images were rendered using ImageJ version 4.3. The images clearly show a high ER colocalization for wt hVKORC1 and the Arg33Ala + Arg35Ala variant. All other variants affecting the Arg98_Arg100 di-arginine motif show a drastic decrease in ER colocalization.
Degree of ER colocalization of hVKORC1 variants. This bar graph represents the comparative mean Pearson’s coefficient and Mander’s R coefficient for wt and mutated hVKORC1 variants affecting the di-arginine and the di-lysine motifs analyzed in this study. Error bars represent the SD. The mean Pearson’s coefficient (gray bars) and Mander’s R (black bars) has been calculated from n = 10 to 24 ROIs. All calculations for Pearson’s and Mander’s R coefficient have been performed on the Image J version 4.3 visualization and analysis software. All hVKORC1 variants except Arg33Ala + Arg35Ala show a significant decrease in ER colocalization compared with the wt.
Degree of ER colocalization of hVKORC1 variants. This bar graph represents the comparative mean Pearson’s coefficient and Mander’s R coefficient for wt and mutated hVKORC1 variants affecting the di-arginine and the di-lysine motifs analyzed in this study. Error bars represent the SD. The mean Pearson’s coefficient (gray bars) and Mander’s R (black bars) has been calculated from n = 10 to 24 ROIs. All calculations for Pearson’s and Mander’s R coefficient have been performed on the Image J version 4.3 visualization and analysis software. All hVKORC1 variants except Arg33Ala + Arg35Ala show a significant decrease in ER colocalization compared with the wt.
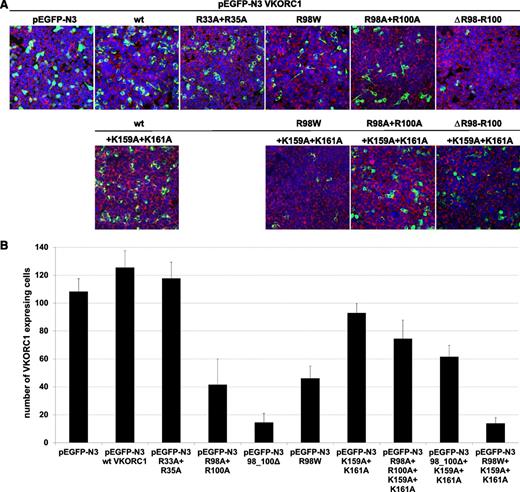
Expression of wt hVKORC1, the mutated di-arginine, and the mutated di-lysine variants. (A) Representative merged overview images for all di-arginine and di-lysine hVKORC1 variants investigated (×40). Human VKORC1 protein is EGFP tagged and green colored, the ER is red fluorescent stained (primary antibody: anti-PDI; secondary antibody: AlexaFluor 594), and the nucleus is counterstained with ToPro3 (blue). (B) The total number of cells expressing the respective hVKORC1 variant per view (×40). Five images of 3 different transfections for each variant were counted. The ratio was normalized to the defined number of 500 cells. Error bars represent the SD.
Expression of wt hVKORC1, the mutated di-arginine, and the mutated di-lysine variants. (A) Representative merged overview images for all di-arginine and di-lysine hVKORC1 variants investigated (×40). Human VKORC1 protein is EGFP tagged and green colored, the ER is red fluorescent stained (primary antibody: anti-PDI; secondary antibody: AlexaFluor 594), and the nucleus is counterstained with ToPro3 (blue). (B) The total number of cells expressing the respective hVKORC1 variant per view (×40). Five images of 3 different transfections for each variant were counted. The ratio was normalized to the defined number of 500 cells. Error bars represent the SD.
Mutation of the arginines belonging to the highly putative di-arginine motif Arg98_Arg100, to alanine, reduced the number of cells with visibly expressed EGFP-tagged protein (n = 42 of 500 cells, Figure 4). The VKCFD2 causing mutation p.Arg98Trp shows similar reduced expression as for the Arg98Ala + Arg100Ala variant (n = 46 of 500 cells, Figure 4). However, for the deletion variant (98_100delRTR), only n = 15 cells were observed to express EGFP-tagged hVKORC1 protein per view (Figure 4). Correlated with the decrease of expression, a characteristic mislocalization of protein was observed for all hVKORC1 variants affecting Arg98_Arg100 (Figures 2-4). A large proportion of the mutated variants were observed not to colocalize with the ER. This effect was most extreme for the deletion variant (98_100delRTR). Evaluation of the degree of colocalization was consistent with the visual observations (Figures 2 and 3). In fact, when the Pearson’s coefficient is taken to indicate the degree of ER colocalization, a significantly lower amount (P < .001) representing ∼8% of the protein (mean Pearson’s coefficient: 0.067 ± 0.035) was colocalized with the ER for the deletion variant compared with the wt (mean Pearson’s coefficient: 0.860 ± 0.062 considered as 100%; Figure 3). The Arg98Ala + Arg100Ala compound substitution and the p.Arg98Trp variant showed ∼15% and 20% ER colocalization, respectively (mean Pearson’s coefficients: 0.130 ± 0.028 and 0.173 ± 0.062, respectively, P < .001 for both; Figure 3). Thus, the mislocalization of all variants corresponding to the Arg98_Arg100 sequence demonstrates its critical importance to ER colocalization.
Mutagenesis of Arg33 and Arg35 to alanine of a second but less putative di-arginine motif showed with n = 118 of 500 cells similar expression to wt hVKORC1 and EGFP (Figure 4). Additionally, we found for this variant an exclusive ER colocalization as high as for wt hVKORC1, indicating that both arginines are not involved in ER retention or retrieval (Figures 2 and 3).
Mutating the lysines to alanines in the hVKORC1 di-lysine motif also had an adverse effect on ER colocalization, although the effect was not as severe as for the Arg98_Arg100 affecting variants. The di-lysine variant Lys159Ala + Lys161Ala resulted in ∼40% ER colocalization with respect to the wt (mean Pearson’s coefficient: 0.348 ± 0.126; Figures 3 and 5). When both the di-lysine and di-arginine motifs were mutated to substitute alanines in a single construct (Arg98Ala + Arg100Ala + Lys159Ala + Lys161Ala), the degree of ER colocalization was only slightly lower than the single substituted di-arginine variant (ie, 14%; mean Pearson’s coefficient: 0.119 ± 0.023; Figures 3 and 5). The combination of the p.Arg98Trp variant and the mutated di-lysine motif showed ER colocalization levels slightly lower than the p.Arg98Trp variant alone (18%; mean Pearson’s coefficient: 0.155 ± 0.020; Figure 3). When the mutated lysines were combined with the deletion variant (Lys159Ala + Lys161Ala + 98_100delRTR), the effect on ER colocalization was the severest (6%; mean Pearson’s coefficient: 0.052 ± 0.020). In addition to the 2 lysines in the di-lysine motif, we also mutated the other charged residues in the adjacent primary sequence (ie, arginine and histidine to alanine, Arg162Ala + His163Ala). Although the variant corresponding to this residue also showed mislocalization, it was not to the extent of the mutated lysines, showing that the lysine residues in the di-lysine motif are more important for ER colocalization than the arginine and histidine (data not shown).
Colocalization images for mutated di-lysine variants in combination with mutated di-arginine motif variants in HEK293T cells. This figure shows representative colocalization images of single immunostained HEK293T cells expressing the Lys159Ala + Lys161Ala, Lys159Ala + Lys161Ala + Arg98Ala + Arg100Ala, Lys159Ala + Lys161Ala + 98_100delRTR, and the Lys159Ala + Lys161Ala + p.Arg98Trp mutant hVKORC1 variants, respectively. Each panel is composed of 4 images; the first one is a merged picture of the green-colored hVKORC1 variant (EGFP tagged), the red-stained ER (primary antibody: anti-PDI; secondary antibody: AlexaFluor 594), and the nucleus counterstained with ToPro3 (blue). Colocalization is represented with increasing intensity of yellow. The second image shows cells expressing the EGFP-tagged VKORC1 protein only, followed by the third image showing the red-stained ER. The last column shows colocalized regions as white dots using the colocalization highlighter plug-in embedded in ImageJ 4.3. All images were rendered using ImageJ version 4.3. The di-lysine–mutated hVKORC1 variant Lys159Ala + Lys161Ala shows a higher amount of ER retention than the di-arginine variants shown in Figure 2. Also, in combination with the di-arginine variants the di-lysine variants do not show a more drastic decrease in ER retention.
Colocalization images for mutated di-lysine variants in combination with mutated di-arginine motif variants in HEK293T cells. This figure shows representative colocalization images of single immunostained HEK293T cells expressing the Lys159Ala + Lys161Ala, Lys159Ala + Lys161Ala + Arg98Ala + Arg100Ala, Lys159Ala + Lys161Ala + 98_100delRTR, and the Lys159Ala + Lys161Ala + p.Arg98Trp mutant hVKORC1 variants, respectively. Each panel is composed of 4 images; the first one is a merged picture of the green-colored hVKORC1 variant (EGFP tagged), the red-stained ER (primary antibody: anti-PDI; secondary antibody: AlexaFluor 594), and the nucleus counterstained with ToPro3 (blue). Colocalization is represented with increasing intensity of yellow. The second image shows cells expressing the EGFP-tagged VKORC1 protein only, followed by the third image showing the red-stained ER. The last column shows colocalized regions as white dots using the colocalization highlighter plug-in embedded in ImageJ 4.3. All images were rendered using ImageJ version 4.3. The di-lysine–mutated hVKORC1 variant Lys159Ala + Lys161Ala shows a higher amount of ER retention than the di-arginine variants shown in Figure 2. Also, in combination with the di-arginine variants the di-lysine variants do not show a more drastic decrease in ER retention.
Colocalization analysis of the plasma membrane and Golgi apparatus with wt and engineered di-arginine and di-lysine motif hVKORC1 variants in HEK293 cells
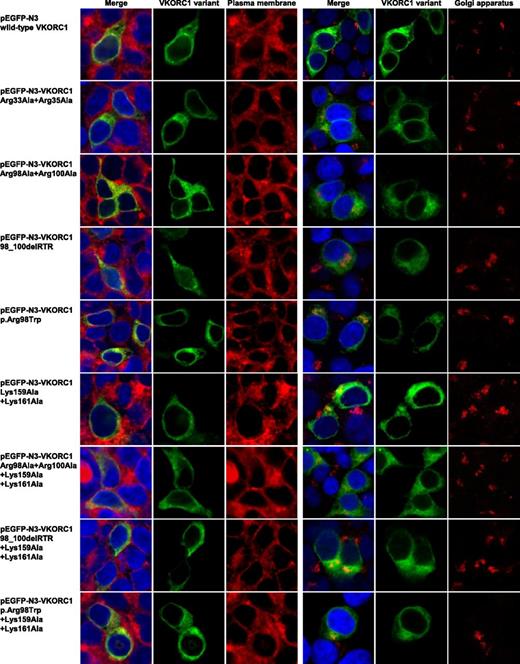
To further investigate the probable final destination of the mislocalized mutated hVKORC1 variants, we stained the plasma membrane as well as the Golgi apparatus. No colocalization was observed for any of the variants or the wt with the Golgi apparatus or the plasma membrane (Figure 6). All mutated variants visually appear to be distributed uniformly across the whole cytoplasm (Figures 2 and 5).
Plasma membrane and Golgi apparatus colocalization images for mutated hVKORC1 di-arginine and di-lysine variants in HEK293T cells. This figure shows colocalization images for single HEK293T cells of immunostained plasma membrane and Golgi apparatus for all hVKORC1 variants investigated in this study. The left column shows merged images in which the nucleus is stained by ToPro3 (blue), the respective EGFP-tagged hVKORC1 variant (green), and the plasma membrane stained by CellMask orange in red. The merged images are followed by images demonstrating the hVKORC1 variant and stained plasma membrane only. On the right side, there are also merged images of the nucleus and the hVKORC1 variant demonstrated in the same way and colors as for the left side. The red fluorescent signal shows the stained Golgi apparatus (anti Golgin-97, AlexaFluor594). The merged images are followed again by images showing the hVKORC1 variant and the stained Golgi apparatus only. No colocalization with the plasma membrane as well as with the Golgi apparatus was found for all hVKORC1 variants investigated including the wt.
Plasma membrane and Golgi apparatus colocalization images for mutated hVKORC1 di-arginine and di-lysine variants in HEK293T cells. This figure shows colocalization images for single HEK293T cells of immunostained plasma membrane and Golgi apparatus for all hVKORC1 variants investigated in this study. The left column shows merged images in which the nucleus is stained by ToPro3 (blue), the respective EGFP-tagged hVKORC1 variant (green), and the plasma membrane stained by CellMask orange in red. The merged images are followed by images demonstrating the hVKORC1 variant and stained plasma membrane only. On the right side, there are also merged images of the nucleus and the hVKORC1 variant demonstrated in the same way and colors as for the left side. The red fluorescent signal shows the stained Golgi apparatus (anti Golgin-97, AlexaFluor594). The merged images are followed again by images showing the hVKORC1 variant and the stained Golgi apparatus only. No colocalization with the plasma membrane as well as with the Golgi apparatus was found for all hVKORC1 variants investigated including the wt.
Discussion
The p.Arg98Trp missense mutation in hVKORC1 is the only one reported so far that results in VKCFD2 in 3 unrelated families.1,5 In an attempt to define the underlying pathophysiological mechanism for this mutation, we have uncovered 2 putative ER retention motifs for hVKORC1, a C-terminal di-lysine motif (KAKRH) and a di-arginine motif at the Arg98_Arg100 position (LRTR or RTRW or RTR). A third candidate di-arginine motif at position Arg33_Arg35 was observed to have no effect on ER retention and therefore does not constitute an ER retention signal.
The putative di-arginine motif at Arg98_Arg100 accounts for the maximum retention of the hVKORC1 protein in the ER, whereas the di-lysine motif appears to have lesser influence. However, both motifs in combination contribute to the complete ER retention of hVKORC1. Interestingly, the human pathogenic variant p.Arg98Trp lies within the di-arginine motif showing also drastic decrease in ER colocalization. Di-arginine ER localization motifs are involved in intracellular sorting and postsynthetic transport of multimeric membrane proteins. In contrast to the C-terminal di-lysine ER retention motif, di-arginine motifs do not need to be exposed at the C terminus of a membrane protein in order to be functional.19 Consistently, naturally occurring di-arginine ER retention motifs are present in various cytosolic domains of polytopic membrane proteins.19 The commonly observed biophysical characteristics of di-arginine ER retention motifs fit very well to the retention motif Arg98_Arg100 that we have identified in hVKORC1, especially when we consider our homology-based membrane-embedded model of hVKORC1.11,19 This model is based on a bacterial homolog of hVKORC1 and consists of 4 transmembrane (4TM) α-helices.10,11 Currently, the membrane topology of hVKORC1 is not fully resolved, and 3TM or 4TM α-helices have been proposed for hVKORC1.10,21-25 However, for both topology models, the putative di-arginine ER retention motif Arg98_Arg100 is predicted to be exposed to the cytoplasmic face of the ER membrane. In the 3TM model, this di-arginine motif is part of the end of the big cytoplasmatic loop close to the second transmembrane α-helix.25 In the 4TM model, the di-arginine motif is located in the short cytoplasmic loop connecting the second and third transmembrane α-helix.10,11 Therefore, both models support the existence of the di-arginine ER retention motif at position Arg98_Arg100. In addition, our data demonstrates that the Arg33_Arg35 sequence has no influence on ER retention, which is in agreement with the 4TM model of hVKORC1, where Arg33_Arg35 is located in the ER lumenal loop (Figures 2 and 3).
Another commonly observed property of di-arginine motifs is their proximity to membrane surfaces (∼15 Å).19 Notably, our homology model indicates that the di-arginine motif Arg98_Arg100 is predicted to lie at the cytoplasmic face of the ER membrane, consistent with the locations of other di-arginine motifs revealed by our in silico motif search (Figure 2; supplemental Table 2). In general, di-arginine motifs are present in different membrane protein families (eg, Kir6.1/2 or SUR1/2 KATP channel subunits) and are often located in poorly conserved or semiconserved regions of the proteins.19 Interestingly, the hVKORC1 region containing the di-arginine motif is semiconserved across all animal species (supplemental Figure 2). The precise mechanisms by which the di-arginine motifs act have been discussed in the literature, but no concrete data identifying specific receptor proteins have been published. However, 2 separate mechanisms are thought to affect ER membrane protein retention in this context. One suggests a direct retention likely involving transmembrane sequences of retained ER proteins, whereas the other involves retrograde transport of membrane proteins that can exit the ER and move into the Golgi compartment before being retrieved to the ER by retrieval proteins such as the coat protein (COP) complex I.19 Neither general principles nor details of the recognition process in the ER retrieval system involving di-arginine motif sequences are currently understood. However, site-directed mutagenesis experiments suggest that the size and/or charge/polarity of the residues comprising this motif might play a role in the recognition process.26 Positively charged arginine residues contribute to the charge- and size-recognition of this motif, whereas the residue after or before the arginine is typically a nonpolar/hydrophobic residue. This is observed for the di-arginine motif of hVKORC1 because the residues after as well as before the arginines are nonpolar (supplemental Table 2). Among the common di-arginine ER retention motifs reported in the literature, the arginine cluster is preceded by a nonpolar residue, usually leucine, a trend that is also observed in the motif in hVKORC1 (supplemental Figure 2).
To further investigate the partial retention of hVKORC1 for the di-arginine motif variants, we searched for alternative signs of ER retention mechanisms and identified an additional di-lysine ER retention motif at the C terminus of hVKORC1. We observed that mutating the di-lysine motif did not have a drastic effect on ER colocalization as observed for the di-arginine motif variants (Figures 2, 3, and 5). The reduction to 40%, with respect to the wt, was about half the effect seen for the di-arginine motif variants. However, mutating both motifs simultaneously resulted in 13% ER colocalization (Arg98Ala + Arg100Ala + Lys159Ala + Lys161Ala; Figure 3). Therefore, our results are consistent with the retention of hVKORC1 inside the ER being primarily mediated by the presence of a wt di-arginine motif. The C-terminal di-lysine motif appears to only augment the di-arginine motif effect slightly. Zerangue et al, while studying ER retrieval signals in mammalians, used a combinatorial approach to generate a series of di-lysine motifs in order to study their efficiency in the trafficking of reporter molecules bearing these C-terminal sequences.26 Their results suggest that C-terminal XKXKXX or KXKXX sequences are not strong ER retention signals. Our results are consistent with their study results. In addition, they observed that in the presence of both motifs the influence on trafficking varies across a wide spectrum depending on the sequence context of each motif, which holds true for our mutated variants as well. However, recent affinity studies suggest that KKXX and KXKXX motifs bind with similar specificities to α-COP and β′-COP proteins.27,28
Disruption of the ER retention signal in a number of proteins has been observed to result in delocalization to the Golgi apparatus or plasma membrane.19 To identify the same for the mutated hVKORC1 proteins affecting both ER retention motifs, we stained the plasma membrane and the Golgi apparatus in cells expressing these variants. All variants did not show colocalization in these organelles, thereby confirming our assumption that most of the mutated protein is mislocalized to the cytoplasm from which it is eventually cleared. Therefore, we conclude that the mutated hVKORC1 protein, although not completely retained in the ER, is also not relocated to either the plasma membrane or to the Golgi apparatus. Because the mutated proteins visually appear to be distributed uniformly across the whole cytoplasm and not clumped (in the event they had been relocated to one of the microorganelles like peroxisomes, their distribution would be uneven and concentrated in certain areas), we suspect that the mutated variants are cleared from the cytoplasm itself (Figures 2 and 5).
In addition to the mislocation of the mutated proteins affecting the 2 ER retention motifs, we also observe markedly reduced protein expression compared with the wt for all variants. This effect was even more pronounced for the deletion variant, which further led to rapid disappearance/degradation of the protein inside the cells as suggested by the very low number of cells showing EGFP-tagged hVKORC1 protein signal (Figure 4). Alternative to compromising ER retention, individual or multiple missense mutations in hVKORC1 might cause loss of protein labeling signal in our experiments for a number of reasons. Improperly folded protein would be sequestered by the ER-associated degradation pathway and degraded, resulting in a loss of protein signal. Proper cotranslational insertion of polytopic membrane proteins such as hVKORC1 requires proper recognition of transmembrane α-helices and surrounding amino acids and sequence motifs. Positively charged residues are known to be important for translocon recognition according to von Heijne’s “positive inside” rule.29 Elimination of Arg98 and Arg100 would disturb the relative number of positively charged residues on both sides of the ER membrane, possibly leading to either misfolding or incomplete insertion into the lipid bilayer. Ultimately, both processes lead to retrotranslocation of unstable proteins into the cytoplasm for proteosomal degradation. Interestingly, 8% to 20% of the RXR variant proteins were retained in the ER relative to wt controls. This suggests that the ER retention function of the di-arginine motif might be complimented by an additional ER retention mechanism. This would be consistent with the fact that the VKCFD2 phenotype of the patients carrying the p.Arg98Trp mutation can be corrected by daily vitamin K supplementation. Supplemented vitamin K might be reduced to KH2 by the relatively low amount of ER-retained VKORC1:p.Arg98Trp to adequately enable sufficient γ-carboxylation to activate sufficient levels of VKCFD for physiological clotting. However, this hypothesis requires direct in vivo and in vitro testing.
A study comparing di-lysine motifs with di-arginine motifs suggested that these 2 signals can be divided into proximal and distal acting signals.30 The di-lysine motif acts best when it is closer to the membrane surface, whereas the di-arginine motif acts best when it is farther from the membrane surface. In our membrane-embedded model, it appears that the di-lysine motif is farther from the membrane surface than the di-arginine motif, which could explain its weaker influence. It is known that the COP complexes interact with the di-lysine motif approximately <20 Å from the transmembrane domains, which further supports the claim.19 Also, we have observed in our 4TM membrane-embedded hVKORC1 model that both these motifs are quite proximal to each other (Figure 1). It can be hypothesized that the interaction with the retrieval COP complexes might actually involve the simultaneous molecular recognition of both motifs to provide a higher degree of specificity as well as binding affinity.
To conclude, we have identified 2 putative motifs important for physiological ER retention of human VKORC1: one is a di-arginine motif (Arg98_Arg100), and the other is a di-lysine motif (Lys159_His163). We have also demonstrated that the hVKORC1 p.Arg98Trp variant disrupts the di-arginine motif resulting in decreased ER retention capacity, subcellular mislocalization, and subsequent degradation of the mutated protein. This leads to a lack of KH2 to drive sufficient γ-carboxylation of VKCFD, thereby resulting in the VKCFD2 phenotype.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported, in part, by funding from Baxter Germany GmbH (J.O.) and by a grant from the Deutsche Forschungsgemeinschaft (Ol100 5-1) (M.W. and J.O.).
Authorship
Contribution: A.B., K.J.C., and J.O. were responsible for experimental design; K.J.C. collected data; K.J.C. and A.B. analyzed data and produced figures; A.B. performed protein modeling; and K.J.C., A.B., S.R., M.W., and J.O. drafted and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Prof Johannes Oldenburg, Institute of Experimental Haematology and Transfusion Medicine, University Clinic Bonn, Sigmund-Freud Strasse 25, 53105 Bonn, Germany; e-mail: johannes.oldenburg@ukb.uni-bonn.de.
References
Author notes
K.J.C. and A.B. contributed equally to this study.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal