Key Points
C/EBPε acetylation regulates C/EBPε transcriptional activity.
C/EBPε acetylation is required for neutrophil differentiation and the formation of neutrophil-specific granules.
Abstract
C/EBPε, a member of the CCAAT/enhancer binding protein (C/EBP) family of transcription factors, is exclusively expressed in myeloid cells and regulates transition from the promyelocytic stage to the myelocytic stage of neutrophil development, being indispensable for secondary and tertiary granule formation. Knowledge concerning the functional role of C/EBPε posttranslational modifications is limited to studies concerning phosphorylation and sumoylation. In the current study, using ectopic expression and ex vivo differentiation of CD34+ hematopoietic progenitor cells, we demonstrate that C/EBPε is acetylated, which was confirmed by mass spectrometry analysis, identifying 4 acetylated lysines in 3 distinct functional domains. Regulation of C/EBPε acetylation levels by the p300 acetyltransferase and the sirtuin 1 deacetylase controls transcriptional activity, which can at least in part be explained by modulation of DNA binding. During neutrophil development, acetylation of lysines 121 and 198 were found to be crucial for terminal neutrophil differentiation and the expression of neutrophil-specific granule proteins, including lactoferrin and collagenase. Taken together, our data illustrate a critical role for acetylation in the functional regulation of C/EBPε activity during terminal neutrophil development.
Introduction
Myeloid differentiation is a closely regulated process, involving differentiation of common myeloid progenitors toward cells of both the granulocyte/macrophage lineage (GM) as well as the megakaryocyte/erythroid lineage.1,2 The regulation of lineage commitment and differentiation has historically been explained by 2 models: signaling by lineage-specific hematopoietic cytokines (deterministic model) and a stochastic model, based on the activity of lineage-specific transcription factors.3-7 With regard to GM lineage progression and determination, CCAAT/enhancer binding protein (C/EBP) factors play an essential role. This is best illustrated in C/EBPα-deficient mice by reduced formation of neutrophils resulting from a defect at the level of the granulocyte/macrophage progenitor.8 Whereas C/EBPα is essential for instruction of the granulocyte/macrophage progenitor towards differentiation of the granulocyte and the monocyte/macrophage lineage, C/EBPε is primarily involved in terminal granulocyte differentiation for both neutrophils and eosinophils.9-12 C/EBPε is the most recently identified member of the C/EBP family of transcription factors and is exclusively expressed in myeloid cells.13,14 During neutrophil development, C/EBPε regulates transition from the promyelocytic stage to the myelocytic stage of differentiation and is essential for secondary and tertiary granule formation.14-16 C/EBPε deficiency, demonstrated in knockout murine models, resembles the clinical phenotype of patients with neutrophil-specific granule deficiency, characterized by an increase in circulating immature neutrophils and recurrent pyogenic infections.12,17-19 As a result of differential RNA splicing and alternative translational start sites, human-C/EBPε is expressed as 4 protein isoforms (32, 30, 27, and 14 kDa) that represent functionally different roles during myeloid differentiation. These functional differences, transcriptional activation by C/EBPε32/30, transcriptional repression by C/EBPε27, and dominant negative regulation by C/EBPε14, can, at least partly, be explained by analysis of the functional domains of human-C/EBPε.20-23
In addition to using activation and repression domains to regulate transcriptional activity, the expression and function of transcription factors is regulated by posttranslational modifications. Concerning C/EBP transcription factors, several posttranslational modification sites have been characterized, primarily on C/EBPα and C/EBPβ.24 Acetylation of C/EBPβ on multiple lysines has been linked functionally to transcriptional activity in previous studies.25-27 Until recently, knowledge of C/EBPε posttranslational modifications was limited to studies concerning phosphorylation and sumoylation. Phosphorylation of C/EBPε on threonine 75, located in the transactivation domain, is associated with increased DNA binding and transcriptional activation by C/EBPε.28,29 Sumoylation of lysine residues within the repression domain has been linked to a conserved repression motif of C/EBPε, similar to other C/EBP family members.30,31 Recently, Kyme et al have demonstrated that C/EBPε can be acetylated in mature peripheral blood neutrophils.32 Here we demonstrate that C/EBPε acetylation is required for neutrophil development. Moreover, we demonstrate that acetylation at lysine 121 is important for C/EBPε DNA binding, and acetylation of both lysines 121 and 198 is indispensable for C/EBPε transcriptional activity during neutrophil development. Taken together, our data provide novel insights into the functional regulation of C/EBPε activity during normal and aberrant neutrophil development and increase the knowledge concerning the functional effects of lysine acetylation on protein function more generally.
Methods
Ethics statement
Umbilical cord blood (UCB) was collected after written informed consent was provided according to the principles expressed in the Declaration of Helsinki. Protocols were approved by the institutional review board of the University Medical Center Utrecht.
Patient samples
Bone marrow specimens were collected during a yearly control visit and bone marrow examination. Written informed consent was obtained from all parents.
Cell lines
COS, HL60, and NB4 cells were maintained according to standard protocols as described in the supplemental Data, available on the Blood Web site.
Antibodies, DNA constructs, and reagents
Isolation, culture, and transduction of human CD34+ cells
The procedures used to isolate and differentiate human CD34+ cells have been described previously.35 Bicistronic retroviral DNA constructs were used, expressing the gene of interest (C/EBPε, C/EBPε K121/198R, C/EBPε R121/198K, and C/EBPε K15xR) and an internal ribosomal entry site, followed by the gene encoding for green fluorescent protein (eGFP). Retroviral transductions were performed as previously described.36
Transfection of cells and luciferase assays
Luciferase assays were performed according to standard protocols, as described in the supplemental Data.
Immunoprecipitation and western blotting
COS cells were cultured and transfected, followed by Flag immunoprecipitation or HA immunoprecipitation, as described previously.37 For immunoprecipitation of endogenous C/EBPε, CD34+ cells were differentiated toward neutrophils over the course of 10 days. Cells were lysed in radioimmunoprecipitation assay lysis buffer, incubated with anti-C/EBPε antibody overnight at 4°C, and incubated with protein A agarose beads (Invitrogen, Life Technologies, Bleiswijk, The Netherlands) the next day. Equal amounts of sample were analyzed by western blot, probed with the respective antibodies. Immunocomplexes were detected using enhanced chemiluminescence (GE Healthcare, Zeist, The Netherlands) or with quantitative fluorescence technology (Odyssey imager, LI-COR biotechnology, Hamburg, Germany).
Confocal studies and proximity ligation assay
The procedures used for confocal analysis have been described previously.37 For proximity ligation assay, COS cells or UCB-derived neutrophils were fixed, blocked, and incubated with primary anti-rabbit and anti-mouse antibody overnight. According to the manufacturer’s protocol (Olink Bioscience, Uppsala, Sweden), cells were subsequently incubated with the secondary mouse PLUS and rabbit MINUS probe for 1 hour at 37°C in a dark humidity chamber. Cells were washed 3 times in proximity ligation assay (PLA) buffer A [10 mM tris(hydroxymethyl)aminomethane-HCl at pH 7.5, 150 mM NaCl, 0.05% Tween in ddH20], followed by ligation for 30 minutes. Again, cells were washed, followed by amplification and detection for 100 minutes at 37°C in a dark humidity chamber. After this, cells were washed 3 times in PLA buffer B [200 mM tris(hydroxymethyl)aminomethane-HCl at pH 7.5, 100 mM NaCl in ddH20] and 3 times in 1% buffer B. Dry cells were mounted in Mowiol 4-88 containing 4,6 diamidino-2-phenylindole. Cells were analyzed with a ×63 objective on a Zeiss LSM 710 fluorescence microscope (Carl Zeiss, Oberkochen, Germany).
Mass spectrometry analysis
Mass spectrometry was performed according to standard protocols, as described in the supplemental Data.
Histochemical and lactoferrin staining of hematopoietic cells
The procedures used to analyze myeloid differentiation have been described previously.34
DNA binding assay
The used procedures for nuclear fractionation are described in the supplemental Data. Double-stranded biotinylated oligos encoding a CEBPε-specific or control DNA binding sequence were coupled to streptavidin agarose beads. Subsequently, oligo-coupled beads were incubated with nuclear fractions isolated from HEK293 cells expressing mutant Flag-CEBPε constructs in binding buffer (10 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, 10 mM KCl, 0,1 mM EGTA, 80 mM NaCl, 1% NP40, 2 mM dithiothreitol, 1% Halt protease inhibitor cocktail). After incubation, beads were washed 5 times with phosphate-buffered saline containing 1% Halt protease inhibitor cocktail and denatured in sodium dodecyl sulfate (SDS) sample buffer before being run on an SDS-acrylamide gel. Separated proteins were then analyzed by western blotting, using Flag-M2 antibody.
Quantitative polymerase chain reaction
Quantitative polymerase chain reaction experiments were performed as previously described.38 Primers are listed in supplemental Table 1. To quantify the data, the comparative threshold cycle method was used. Relative quantity was defined as 2−ΔΔCt. β2M was used as the reference gene.
Statistical analysis
Statistical analysis was performed using a one-way analysis of variance test, followed by a Bonferroni multiple comparison test (comparison between all conditions) or a Dunnet multiple comparison test (comparison with the control) (Prism GraphPad Software). P values of .05 or less were considered significant (*P < .05; **P < .01).
Results
C/EBPε is acetylated in neutrophils
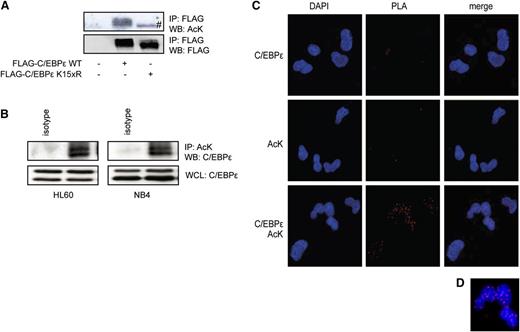
To investigate whether C/EBPε is acetylated, cells were transfected with an epitope-tagged C/EBPε construct and acetylation was analyzed by immunoprecipitation and immunoblotting, using an anti-acetyl lysine antibody (anti-AcK). A high basal level of C/EBPε acetylation was observed (Figure 1A). To validate the specificity of the anti-acetyl lysine antibody, we constructed a lysine dead mutant of C/EBPε in which all 15 lysines of C/EBPε were mutated into arginine residues. Mutating all lysines did not affect protein expression or stability, suggesting this mutant protein is properly folded (supplemental Figure 1). No acetylation of the lysine dead mutant was observed; the lower band was considered nonspecific (Figure 1A). To determine whether endogenously expressed C/EBPε was also acetylated, we analyzed the myeloid cell lines HL60 and NB4. NB4 cells were pretreated with retinoic acid for 4 days to increase C/EBPε expression.39 Protein lysates were prepared, followed by acetyl lysine-immunoprecipitation and immunoblotting, using a C/EBPε antibody. Compared with isotype control, we observed C/EBPε acetylation of the 32/30 kD isoforms (Figure 1B), suggesting that C/EBPε is acetylated in these myeloid cell lines. In addition, CD34+ cells from UCB were differentiated toward neutrophils for 10 days, followed by analysis of C/EBPε acetylation utilizing PLA (Methods). Given that a PLA signal can only be obtained when detection antibodies are in extremely close proximity, this technique enables the detection of posttranslational modifications in their cellular context. At day 10 of neutrophil differentiation, cytospins were prepared and cells were incubated with an anti-acetyl lysine antibody and anti-C/EBPε antibody. After PLA, we observed a specific nuclear signal only on incubation with both antibodies. Although there is a theoretical possibility that this reflects the interaction of C/EBPε with another acetylated protein, these data suggest that C/EBPε is acetylated in neutrophil progenitor cells (Figures 1C-D).
C/EBPε is acetylated in neutrophil progenitors. (A) COS cells were transfected with Flag-C/EBPε or Flag-C/EBPε K15xR, followed by Flag immunoprecipitation. Cell lysates were analyzed by WB, using anti-acetylated lysines (AcK) and anti-Flag antibodies. (B) NB4 cells were treated with retinoic acid over the course of 4 days. HL60 cells and NB4 cells were treated with KDACi over the course of 3 hours, followed by AcK immunoprecipitation. Cell lysates were analyzed by WB, using anti-C/EBPε and AcK antibodies. (C) PLA was performed in CD34+-derived neutrophil progenitors, probed for C/EBPε and AcK. The PLA signal (red dots) represents acetylation of C/EBPε. (D) Magnified view of PLA signal. Data are representative of at least 3 independent experiments. IP, immunoprecipitation; WB, western blot; WCL, whole-cell lysate. #, a specific band.
C/EBPε is acetylated in neutrophil progenitors. (A) COS cells were transfected with Flag-C/EBPε or Flag-C/EBPε K15xR, followed by Flag immunoprecipitation. Cell lysates were analyzed by WB, using anti-acetylated lysines (AcK) and anti-Flag antibodies. (B) NB4 cells were treated with retinoic acid over the course of 4 days. HL60 cells and NB4 cells were treated with KDACi over the course of 3 hours, followed by AcK immunoprecipitation. Cell lysates were analyzed by WB, using anti-C/EBPε and AcK antibodies. (C) PLA was performed in CD34+-derived neutrophil progenitors, probed for C/EBPε and AcK. The PLA signal (red dots) represents acetylation of C/EBPε. (D) Magnified view of PLA signal. Data are representative of at least 3 independent experiments. IP, immunoprecipitation; WB, western blot; WCL, whole-cell lysate. #, a specific band.
Acetylation of C/EBPε is regulated by SIRT1 and p300
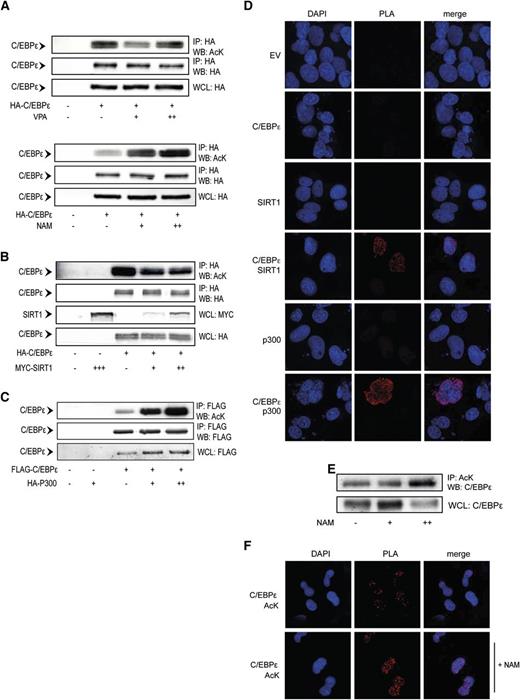
Acetylation is regulated by lysine acetyltransferases and lysine deacetylases (KDAC). To investigate the specific regulators of C/EBPε acetylation, we first investigated the effects of the class I/II KDAC inhibitor valproic acid (VPA) and the sirtuin 1 (SIRT1) inhibitor nicotinamide (NAM) on C/EBPε acetylation. Cells were transfected with epitope-tagged C/EBPε and treated overnight with increasing concentrations of VPA or NAM. C/EBPε was immunoprecipitated, followed by acetyl lysine immunoblotting. Although on treatment with VPA (Figure 2A, upper) we observed no reproducible effects on C/EBPε acetylation, treatment with NAM (Figure 2A, lower) resulted in a concentration-dependent increase in C/EBPε acetylation. This suggests SIRT1 may regulate C/EBPε deacetylation. To further investigate the role of SIRT1 as a regulator of C/EBPε acetylation, we cotransfected SIRT1 with C/EBPε and again performed immunoprecipitation, followed by anti-acetyl lysine immunoblotting. Cotransfection with SIRT1 decreased the levels of C/EBPε acetylation, suggesting SIRT1 deacetylates C/EBPε (Figure 2B).
C/EBPε acetylation is regulated by SIRT1 and p300. (A) COS cells were transfected with HA-C/EBPε and treated overnight in the absence or presence of 500 μM (+), 1 mM (++) VPA, 1 mM (+), 5 mM (++) NAM, followed by HA immunoprecipitation. Cell lysates were analyzed by WB, using anti-acetyl lysines (AcK) and anti-HA antibodies. (B) COS cells were cotransfected with HA-C/EBPε and Myc-SIRT1, followed by HA immunoprecipitation. Cell lysates were analyzed by WB, using anti-AcK, anti-HA, and anti-Myc antibodies. (C) COS cells were cotransfected with Flag-C/EBPε and HA-p300, followed by Flag immunoprecipitation. Cell lysates were analyzed by WB, using anti-AcK, anti-Flag, and anti-HA antibodies. (D) COS cells transfected with FLAG-C/EBPε and HA-p300 or Myc-SIRT1, followed by PLA. Cells were probed for Flag, HA, or Myc. Single transfections and pMT2 empty vector were used as controls. PLA signal (red dots) represents colocalization of C/EBPε with p300 and SIRT1. (E) CD34+ cells were differentiated toward neutrophils. At day 10, cells were treated overnight with 5 mM NAM, followed by AcK immunoprecipitation. Cell lysates were analyzed by WB, using an anti-C/EBPε antibody. (F) Day 10 neutrophil precursors were isolated and treated overnight with NAM, followed by PLA. Cells were probed with anti-AcK and anti-C/EBPε antibodies. PLA signal (red dots) represents C/EBPε acetylation. IP, immunoprecipitation; NAM, nicotinamide; VPA, valproic acid; WB, western blot; WCL, whole-cell lysate. Data are representative of 3 or more independent experiments.
C/EBPε acetylation is regulated by SIRT1 and p300. (A) COS cells were transfected with HA-C/EBPε and treated overnight in the absence or presence of 500 μM (+), 1 mM (++) VPA, 1 mM (+), 5 mM (++) NAM, followed by HA immunoprecipitation. Cell lysates were analyzed by WB, using anti-acetyl lysines (AcK) and anti-HA antibodies. (B) COS cells were cotransfected with HA-C/EBPε and Myc-SIRT1, followed by HA immunoprecipitation. Cell lysates were analyzed by WB, using anti-AcK, anti-HA, and anti-Myc antibodies. (C) COS cells were cotransfected with Flag-C/EBPε and HA-p300, followed by Flag immunoprecipitation. Cell lysates were analyzed by WB, using anti-AcK, anti-Flag, and anti-HA antibodies. (D) COS cells transfected with FLAG-C/EBPε and HA-p300 or Myc-SIRT1, followed by PLA. Cells were probed for Flag, HA, or Myc. Single transfections and pMT2 empty vector were used as controls. PLA signal (red dots) represents colocalization of C/EBPε with p300 and SIRT1. (E) CD34+ cells were differentiated toward neutrophils. At day 10, cells were treated overnight with 5 mM NAM, followed by AcK immunoprecipitation. Cell lysates were analyzed by WB, using an anti-C/EBPε antibody. (F) Day 10 neutrophil precursors were isolated and treated overnight with NAM, followed by PLA. Cells were probed with anti-AcK and anti-C/EBPε antibodies. PLA signal (red dots) represents C/EBPε acetylation. IP, immunoprecipitation; NAM, nicotinamide; VPA, valproic acid; WB, western blot; WCL, whole-cell lysate. Data are representative of 3 or more independent experiments.
Next, we investigated whether the ubiquitously expressed lysine acetyltransferase p300 could acetylate C/EBPε. p300 was transfected together with C/EBPε, followed by immunoprecipitation and immunoblotting, as described before. A concentration-dependent increase of C/EBPε acetylation on cotransfection of p300 was observed, suggesting p300 can mediate C/EBPε acetylation (Figure 2C). To determine whether C/EBPε also interacts with SIRT1 and p300, we investigated association of these proteins, using PLA. We observed specific PLA signal on cotransfection of C/EBPε together with either SIRT1 or p300, but not in single-transfection controls, demonstrating that SIRT1 and p300 both interact with C/EBPε (Figure 2D; supplemental Figure 2). To investigate the role of SIRT1 in the regulation of endogenously expressed C/EBPε, CD34+ cells were differentiated toward neutrophils ex vivo. Cells were treated overnight with nicotinamide, followed by immunoprecipitation and PLA. Compared with control cells, treatment with NAM resulted in increased levels of acetylated C/EBPε, observed by both immunoblotting (Figure 2E) and PLA (Figure 2F; supplemental Figure 3), suggesting SIRT1 can regulate C/EBPε acetylation in human neutrophil precursors. Taken together, these data indicate that C/EBPε can be acetylated by p300 and deacetylated by SIRT1.
Acetylation of C/EBPε regulates transcriptional activity
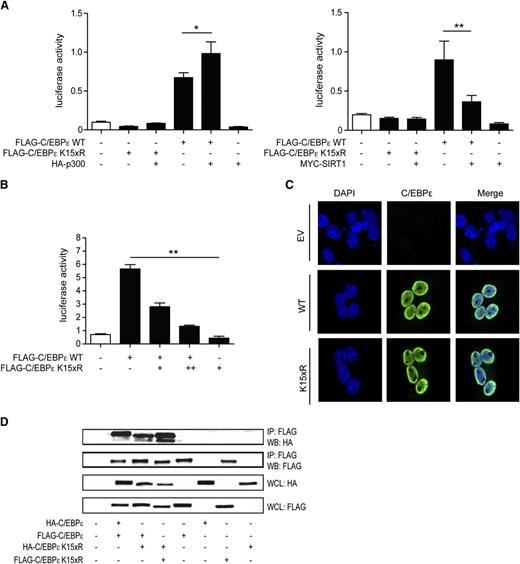
To investigate the functional role of C/EBPε acetylation, luciferase assays were performed using a reporter plasmid containing the macrophage colony-stimulating factor receptor (M-CSFR) promoter; a canonical transcriptional target of C/EBPs.40 Cells were transfected with epitope-tagged C/EBPε or lysine “dead” C/EBPε K15xR in the absence or presence of p300 or SIRT1. Transfection of C/EBPε induced reporter activity, which was further increased on cotransfection of p300 and decreased on cotransfection of SIRT1. Moreover, transfection of C/EBPε K15xR resulted in no transcriptional activation (Figure 3A). In addition C/EBPε was transfected together with an increasing amount of the lysine-dead variant (K15xR), which resulted in a concentration-dependent inhibition of reporter activity (Figure 3B). This suggests that C/EBPε K15xR acts as a dominant-negative transcriptional regulator or prevents binding of C/EBPε to the M-CSFR promotor. Taken together, these data suggest that C/EBPε acetylation is essential for transcriptional activation and that acetylation levels regulate transcriptional activity.
Acetylation of C/EBPε regulates transcriptional activity. (A) M-CSFR promoter luciferase reporter activity was analyzed in COS cells by the cotransfection of C/EBPε or C/EBPε K15xR with p300 or SIRT1. An empty vector plasmid was used as control. All values were normalized for cotransfected renilla. (B) COS cells were cotransfected with C/EBPε and increasing concentrations of K15xR, followed by analysis of C/EBPε-specific luciferase reporter activity. All values were normalized for cotransfected renilla. (C) COS cells were transfected with Flag-C/EBPε or Flag-C/EBPε K15xR, followed by confocal microscopy imaging using anti-Flag antibody and 4,6 diamidino-2-phenylindole nuclear staining. Untransfected cells were used as control. (D) COS cells were cotransfected with Flag- or HA-tagged C/EBP and Flag- or HA-tagged C/EBPε K15xR, followed by FLAG immunoprecipitation. Cell lysates were analyzed by WB, using anti-HA antibody. IP, immunoprecipitation; WB, western blot; WCL, whole-cell lysate. Data are representative of 3 or more independent experiments; error bars represent standard error of the mean (between experiments). *P < .05; **P < .01.
Acetylation of C/EBPε regulates transcriptional activity. (A) M-CSFR promoter luciferase reporter activity was analyzed in COS cells by the cotransfection of C/EBPε or C/EBPε K15xR with p300 or SIRT1. An empty vector plasmid was used as control. All values were normalized for cotransfected renilla. (B) COS cells were cotransfected with C/EBPε and increasing concentrations of K15xR, followed by analysis of C/EBPε-specific luciferase reporter activity. All values were normalized for cotransfected renilla. (C) COS cells were transfected with Flag-C/EBPε or Flag-C/EBPε K15xR, followed by confocal microscopy imaging using anti-Flag antibody and 4,6 diamidino-2-phenylindole nuclear staining. Untransfected cells were used as control. (D) COS cells were cotransfected with Flag- or HA-tagged C/EBP and Flag- or HA-tagged C/EBPε K15xR, followed by FLAG immunoprecipitation. Cell lysates were analyzed by WB, using anti-HA antibody. IP, immunoprecipitation; WB, western blot; WCL, whole-cell lysate. Data are representative of 3 or more independent experiments; error bars represent standard error of the mean (between experiments). *P < .05; **P < .01.
Under homeostatic conditions, CEBPε is exclusively localized in the nucleus. To investigate whether acetylation can regulate nuclear localization of C/EBPε, confocal analysis of cells cotransfected with C/EBPε and C/EBPε K15xR was performed. C/EBPε and C/EBPε K15xR both localized in the nucleus, indicating that C/EBPε acetylation is not critical for nuclear localization (Figure 3C).
C/EBP proteins are characterized by a highly conserved basic leucine zipper domain, which is in involved in homodimerization and heterodimerization of C/EBPs and is essential for DNA binding. To determine whether acetylation is important for the dimerization potential of C/EBPε, coimmunoprecipitations were performed with epitope-tagged C/EBPε and C/EBPε K15xR. We observed no differences in coimmunoprecipitation whether C/EBPε was cotransfected with either C/EBPε or C/EBPε K15xR, and no effect on coimmunoprecipitation of C/EBPε K15xR with C/EBPε K15xR, suggesting acetylation of C/EBPε is also not important for dimerization capacity (Figure 3D). In summary, these data suggest that C/EBPε acetylation modulates transcriptional activity and is not specifically involved in dimerization capacity or nuclear localization.
C/EBPε acetylation levels depend on acetylation of lysine 121 and 198
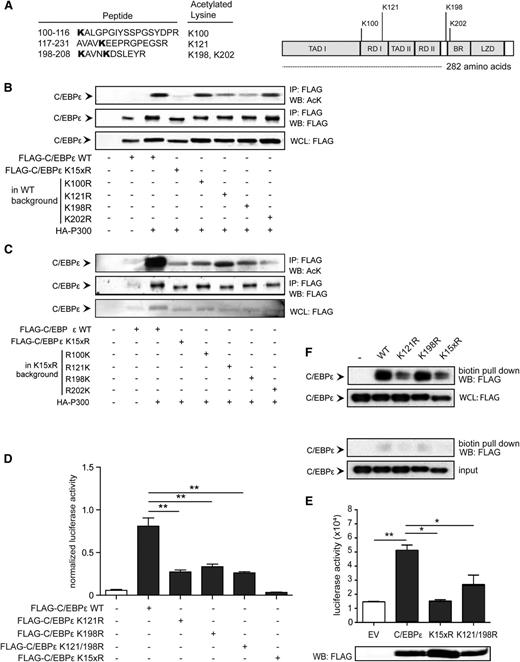
Acetylation is a posttranslational modification that is restricted to lysine residues. C/EBPε (32/30 kDa) contains 15 lysine residues located in all 6 functional domains. To define which lysines can be acetylated, a mass spectrometry analysis was performed. Briefly, C/EBPε was transfected together with p300 and cells were treated with NAM to increase acetylation levels. Lysates were prepared, followed by immunoprecipitation and SDS-polyacrylamide gel electrophoresis. C/EBPε protein bands were isolated and processed for mass spectrometry analysis. We identified 4 acetylated lysines, including 2 lysines in repression domain (RD) I (K100 and K121), 1 lysine between RD II and the basic region (K198), and 1 lysine (K202) in the basic region, containing the DNA binding domain (Figure 4A).
Acetylation of lysine 121 and 198 regulates C/EBPε activity. (A) COS cells were transfected with Flag-C/EBPε together with HA-p300 and treated with NAM. C/EBPε protein bands were isolated from the lysates and processed for mass spectrometry analysis. Data represent the identified lysines and functional domain location. (B-C) COS cells were transfected with Flag-C/EBPε single-lysine knock-out (from WT) and “addback” (from lysine dead) together with HA-p300, followed by Flag immunoprecipitation. Cell lysates were analyzed by WB, using anti-acetylated lysines (AcK) and anti-Flag antibody. (D) M-CSFR promoter luciferase activity was analyzed in COS cells transfected with Flag-C/EBPε, Flag-C/EBPε K121R, Flag-C/EBPε K198R, Flag-C/EBPε K121/198R, and C/EBPε K15xR. An empty vector plasmid was used as control. All values were normalized for cotransfected renilla. (E) Lactoferrin reporter activity was analyzed in COS cells transfected with Flag-C/EBPε, Flag-C/EBPε K121/198R, or C/EBPε K15xR. Cell lysates were analyzed for C/EBPε expression by WB, using an anti-Flag antibody. (F) Nuclear lysates were prepared from COS cells transfected with Flag-C/EBPε, Flag-C/EBPε K121R, Flag-C/EBPε K198R, and Flag-C/EBPε K15xR, followed by pull-down of biotinylated oligonucleotides. Cell lysates were analyzed by WB, using anti-Flag antibody. As controls, oligonucleotides representing a mutated binding site were used. IP, immunoprecipitation; WB, western blot; WCL, whole-cell lysate. Data (B-F) are representative of 3 or more independent experiments; error bars represent standard error of the mean (between experiments). *P < .05; **P < .01.
Acetylation of lysine 121 and 198 regulates C/EBPε activity. (A) COS cells were transfected with Flag-C/EBPε together with HA-p300 and treated with NAM. C/EBPε protein bands were isolated from the lysates and processed for mass spectrometry analysis. Data represent the identified lysines and functional domain location. (B-C) COS cells were transfected with Flag-C/EBPε single-lysine knock-out (from WT) and “addback” (from lysine dead) together with HA-p300, followed by Flag immunoprecipitation. Cell lysates were analyzed by WB, using anti-acetylated lysines (AcK) and anti-Flag antibody. (D) M-CSFR promoter luciferase activity was analyzed in COS cells transfected with Flag-C/EBPε, Flag-C/EBPε K121R, Flag-C/EBPε K198R, Flag-C/EBPε K121/198R, and C/EBPε K15xR. An empty vector plasmid was used as control. All values were normalized for cotransfected renilla. (E) Lactoferrin reporter activity was analyzed in COS cells transfected with Flag-C/EBPε, Flag-C/EBPε K121/198R, or C/EBPε K15xR. Cell lysates were analyzed for C/EBPε expression by WB, using an anti-Flag antibody. (F) Nuclear lysates were prepared from COS cells transfected with Flag-C/EBPε, Flag-C/EBPε K121R, Flag-C/EBPε K198R, and Flag-C/EBPε K15xR, followed by pull-down of biotinylated oligonucleotides. Cell lysates were analyzed by WB, using anti-Flag antibody. As controls, oligonucleotides representing a mutated binding site were used. IP, immunoprecipitation; WB, western blot; WCL, whole-cell lysate. Data (B-F) are representative of 3 or more independent experiments; error bars represent standard error of the mean (between experiments). *P < .05; **P < .01.
To investigate the functional role of these specific lysines, we constructed single-lysine mutants and “addback” mutants in both the wild-type and lysine-dead background, respectively. To determine the role of these lysines in C/EBPε acetylation, we transfected lysine mutant C/EBPε proteins together with p300 and performed immunoprecipitation followed by immunoblotting with the anti-acetyl lysine antibody. Compared with C/EBPε, we observed a clear decrease in total C/EBPε acetylation in the C/EBPε K121R and C/EBPε K198R mutants, although this was not observed in the C/EBPε K100R and C/EBPε K202R mutants (Figure 4B). To confirm these findings, C/EBPε and C/EBPε K15xR or the add-back mutants C/EBPε R100K, C/EBPε R121K, C/EBPε R198K, and C/EBPε R202K were transfected together with p300, followed by immunoprecipitation and immunoblotting. Compared with C/EBPε K15xR, a clear increase in acetylation was observed for C/EBPε R121K, and a less pronounced effect for the C/EBPε R198K mutant (Figure 4C). Taken together, these data suggest that K121 and K198 are important for C/EBPε acetylation.
To investigate whether acetylation of these specific lysines is also functionally relevant, a luciferase assay was performed again, using both M-CSFR and a lactoferrin promoter luciferase reporter, with lactoferrin being a C/EBPε target gene. Compared with reporter activity induced by C/EBPε, a significant decrease in transcriptional activity on transfection of the C/EBPε K121R, C/EBPε K198R, and C/EBPε K121/198R mutants (Figure 4D and Figure 4E) was observed, whereas this was not observed for the C/EBPε K100R and C/EBPε K202R mutants (supplemental Figure 4A), suggesting acetylation of K121 and K198 is important for C/EBPε function. Because C/EBPε acetylation did not affect nuclear localization and dimerization capacity, we next investigated whether C/EBPε acetylation is important for DNA binding. We transfected cells with C/EBPε, C/EBPε K121R, C/EBPε K198R, and C/EBPε K15xR and prepared nuclear lysates. Next, a DNA binding assay was performed using pull-down of biotinylated oligonucleotides representing a specific C/EBPε binding site or control DNA representing a mutated binding site. A decrease in DNA binding was observed for the C/EBPε K121R mutant and K15xR mutant (Figure 4F). In addition, DNA binding could be rescued on transfection of C/EBPε K121Q, representing an “acetylation mimic” (supplemental Figure 4B). Together, this suggests that acetylation of C/EBPε and, more specifically, K121 is important for DNA binding.
Acetylation of C/EBPε K121 and K198 is functionally important during terminal neutrophil differentiation
C/EBPε regulates the transition from the promyelocytic stage into the myelocytic stage of neutrophil development and is essential for the formation of secondary and tertiary granules, including lactoferrin, and collagenase.15,19 We have demonstrated that C/EBPε is acetylated in neutrophils and that acetylation levels can be increased by treatment with the SIRT1 inhibitor NAM. To investigate the functional role of C/EBPε acetylation during neutrophil development, we first investigated the effects of NAM on terminal neutrophil differentiation. Although no increase in the percentage of mature neutrophils was observed, treatment with NAM induced a clear increase in lactoferrin expression (Figure 5A) and the percentage of metamyelocytes (supplemental Figure 5A).
Acetylation of C/EBPε K121 and K198 is functionally important during neutrophil differentiation. CD34+ cells were differentiated toward neutrophils in the absence or presence of 5 mM NAM. (A) In UCB-derived cells, neutrophil differentiation was determined by cytospin analysis of neutrophil morphology and fluorescence-activated cell sorter analysis of intracellular lactoferrin expression (mean fluorescence intensity [MFI]). (B) In SCN patient BM-derived cells, neutrophil development was determined by cytospin analysis of the percentage mature neutrophils and metamyelocytes. (C) At day 7 of differentiation, intracellular lactoferrin expression (MFI) was analyzed and protein lysates were prepared, followed by western blot analysis of C/EBPε expression. (D) CD34+ cells from UCB were retrovirally transduced with eGFP-PMX-C/EBPε, eGFP-PMX-C/EBPε eGFP-PMX-C/EBPε K121/198R, eGFP-PMX-C/EBPε K15xR, and C/EBPε R121/198K and differentiated toward neutrophils. As control, an empty vector was used. Sorted cells were analyzed after 14 days. Neutrophil differentiation was determined by cytospin analysis of morphology and fluorescence-activated cell sorter analysis of lactoferrin expression (MFI). (E) mRNA expression of lactoferrin and collagenase was analyzed by real-time quantitative polymerase chain reaction. Data represent the expression of lactoferrin and collagenase relative to the empty vector. Data are representative of 3 (or more) UCB donors or patient samples. Error bars represent standard error of the mean (between donors). *P < .05; **P < .01.
Acetylation of C/EBPε K121 and K198 is functionally important during neutrophil differentiation. CD34+ cells were differentiated toward neutrophils in the absence or presence of 5 mM NAM. (A) In UCB-derived cells, neutrophil differentiation was determined by cytospin analysis of neutrophil morphology and fluorescence-activated cell sorter analysis of intracellular lactoferrin expression (mean fluorescence intensity [MFI]). (B) In SCN patient BM-derived cells, neutrophil development was determined by cytospin analysis of the percentage mature neutrophils and metamyelocytes. (C) At day 7 of differentiation, intracellular lactoferrin expression (MFI) was analyzed and protein lysates were prepared, followed by western blot analysis of C/EBPε expression. (D) CD34+ cells from UCB were retrovirally transduced with eGFP-PMX-C/EBPε, eGFP-PMX-C/EBPε eGFP-PMX-C/EBPε K121/198R, eGFP-PMX-C/EBPε K15xR, and C/EBPε R121/198K and differentiated toward neutrophils. As control, an empty vector was used. Sorted cells were analyzed after 14 days. Neutrophil differentiation was determined by cytospin analysis of morphology and fluorescence-activated cell sorter analysis of lactoferrin expression (MFI). (E) mRNA expression of lactoferrin and collagenase was analyzed by real-time quantitative polymerase chain reaction. Data represent the expression of lactoferrin and collagenase relative to the empty vector. Data are representative of 3 (or more) UCB donors or patient samples. Error bars represent standard error of the mean (between donors). *P < .05; **P < .01.
Severe congenital neutropenia (SCN) is characterized by the absence of mature neutrophils accompanied by a differentiation block at the promyelocytic stage of differentiation.3 In bone marrow-derived myeloid progenitors from patients with SCN, an increase in the percentage of mature neutrophils, metamyelocytes, was observed on NAM treatment (Figure 5B). In addition, the effects in UCB and BM-derived cells were accompanied by a significant decrease in the percentage of monocytes (supplemental Figure 5B). Because these effects were accompanied by increased lactoferrin expression, with no increase in C/EBPε protein expression (Figure 5C), this suggests that the effects of NAM treatment on neutrophil maturation could at least be partially explained by increased C/EBPε activity.
To demonstrate the functional role of C/EBPε acetylation specifically, UCB CD34+ cells were retrovirally transduced with C/EBPε, C/EBPε K121/198R, C/EBPε K15xR, or C/EBPε R121/198K and were differentiated toward neutrophils. As a control, CD34+ cells transduced with an empty vector were used. On transduction with the K121/198R mutant, we observed a reduction in the percentage of mature neutrophils and intracellular lactoferrin expression (Figure 5D). These effects were enhanced after transduction with the C/EBPε K15xR (lysine dead) mutant. Strikingly, these effects could be almost completely reversed after add-back of lysines 121 and 198, illustrated by an increase in the percentage of mature neutrophils and intracellular lactoferrin expression (Figure 5D). Together, these data suggest acetylation of K121 and K198 of C/EBPε is important for neutrophil differentiation.
To confirm these effects were C/EBPε-specific, we analyzed the effects of the C/EBPε mutants on the expression of specific transcriptional targets, including lactoferrin, and collagenase. Compared with the cells transfected with the empty vector, transduction with the lysine dead mutant abrogated lactoferrin and collagenase expression, whereas the K121 and K198 add-back mutant (R121/198K) rescued expression to approximately baseline levels (Figure 5E).
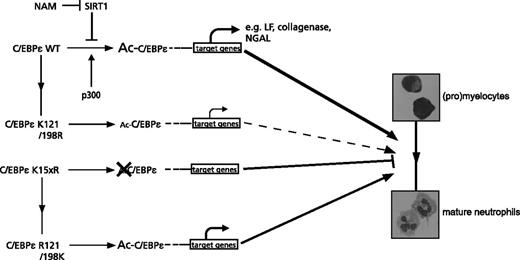
Taken together, these data suggest that during neutrophil development, the regulation of C/EBPε acetylation, predominantly K121 and K198, is important for C/EBPε transcriptional activity (Figure 6).
The role of C/EBPε acetylation for C/EBPε transcriptional activity during neutrophil differentiation. Regulation of C/EBPε acetylation, most importantly lysines 121 and 198, by the lysine acetyltransferase p300 and the lysine deacetylase SIRT1 is involved in the regulation of DNA binding and determines C/EBPε transcriptional activity and the subsequent expression of C/EBPε target genes during neutrophil development. In the absence of C/EBPε acetylation, represented by the lysine dead mutant, C/EBPε activity is abrogated, resulting in a block in neutrophil differentiation.
The role of C/EBPε acetylation for C/EBPε transcriptional activity during neutrophil differentiation. Regulation of C/EBPε acetylation, most importantly lysines 121 and 198, by the lysine acetyltransferase p300 and the lysine deacetylase SIRT1 is involved in the regulation of DNA binding and determines C/EBPε transcriptional activity and the subsequent expression of C/EBPε target genes during neutrophil development. In the absence of C/EBPε acetylation, represented by the lysine dead mutant, C/EBPε activity is abrogated, resulting in a block in neutrophil differentiation.
Discussion
In the present study, we have investigated the functional relevance of C/EBPε acetylation. Our data demonstrate that C/EBPε is acetylated and that this modification is important for neutrophil maturation. We have previously demonstrated that modulating protein acetylation in CD34+ hematopoietic progenitors, by using lysine deacetylase inhibitors (KDACi), affects cell fate decisions during myeloid development.35 These data suggested that the effects were the result of regulation of both histone and nonhistone proteins.
Skokowa et al have demonstrated that treatment of myeloid progenitors with NAM stimulates neutrophil differentiation both in vitro and in vivo.41 Recently, Kyme et al demonstrated that the antibacterial function of both human and murine neutrophils could be increased by treatment with NAM, modulating the expression of C/EBPε, and downstream effectors.32 In addition, and in agreement with our data, they demonstrate that C/EBPε acetylation levels can be increased by treatment with NAM. In contrast to this study, we observed no increase in C/EBPε protein expression, which can be explained by differential C/EBPε expression in immature neutrophil precursors compared with activated neutrophils in the peripheral blood. Furthermore, in our study, we demonstrate for the first time the specific functional role of C/EBPε acetylation. We demonstrate that modulation of C/EBPε acetylation levels by p300 and SIRT1 regulates transcriptional activation by C/EBPε. By mass spectrometry analysis, we identified 4 acetylated lysines, and the functional effects we observed could at least be partly allocated to acetylation of 2 specific lysines: K121 and K198. K121 is located in the repression domain I of C/EBPε, which has previously been identified as a key element of the conserved repression motif (RDM) of various C/EBP family members, including C/EBPβ.21,31 The regulation of C/EBPε transcriptional activity has previously been associated with levels of K121 sumoylation, suggesting a delicate balance between repression and the recruitment of coactivators, resulting in increased C/EBPε transcriptional activity.30,31 According to our data, acetylation of K121 is important for DNA binding and transcriptional activity. This was confirmed by increased C/EBPε activity in luciferase reporter assays and the rescue of DNA binding using a C/EBPε mutant in which lysine 121 was replaced by a glutamine residue (K121Q), mimicking acetylation (supplemental Figures 4C and 6). The balance between K121 acetylation, involved in DNA binding of C/EBPε (Figure 4), and K121 sumoylation, involved in active repression, and recruitment of cofactors could regulate RDM and RDI function and thereby modulate C/EBPε transcriptional activity. Although mutating lysine 198 to arginine, mimicking deacetylation, resulted in a clear decrease in total C/EBPε acetylation and transcriptional activity (Figure 4), the specific functional role of K198 acetylation still requires further investigation. K198 is located in close proximity to the DNA binding region and a highly conserved residue between species and C/EBP family members, including C/EBPα and C/EBPβ. Because K121R and K15xR still retain some DNA binding activity (Figure 4E; supplemental Figure 4C), whereas heterodimerization capacity is unaffected (Figure 3D), this suggests that these lysine mutants, including K198R, potentially regulate protein–protein interactions, resulting in suppression of C/EBPε target genes.
Interestingly, although C/EBPε is indispensable for neutrophil maturation, overexpression of C/EBPε in CD34+ cells did not increase the percentage of mature neutrophils and the expression of C/EBPε transcriptional targets (Figure 5). Bedi et al previously demonstrated that enforced expression of the transcriptional activator isoforms C/EBPε32/30 in hematopoietic progenitor cells stimulated eosinophil differentiation at the expense of granulocyte/macrophage differentiation, whereas overexpression of the C/EBPε27 or C/EBPε14 isoforms inhibited eosinophil differentiation.20 In agreement with these data, we observed disturbed neutrophil maturation accompanied by an increase in eosinophil precursors in some, but not all, donors, suggesting this is dependent on C/EBPε32/30 expression levels. Intriguingly, compared with the lysine dead mutant, enforced expression of C/EBPε R121/198K rescued neutrophil differentiation, yet induced no increase in eosinophil precursors or aberrant neutrophil maturation, suggesting these lysines are selectively important for terminal neutrophil differentiation (supplemental Figure 7). In this study, we have only investigated the functional role of acetylation of full-length C/EBPε during neutrophil differentiation. Whether acetylation of K121 and K198R, located in domains that are identical in all 4 isoforms of C/EBPε (32, 30, 27, and 14 kDa), differentially regulates these C/EBPε isoforms functionally during myeloid differentiation requires further investigation.
Terminal neutrophil differentiation is the result of a complex interplay between transcription factors and modulators, including PU.1, CCAAT displacement protein (CDP), the retinoic acid receptor (RAR), and C/EBPε.9,42,43 Studies in C/EBPε-deficient mice have demonstrated that this transcription factor is indispensible for neutrophil maturation and function.12,44 Moreover, loss of C/EBPε function abrogates the production of specific neutrophil granule proteins, resulting in disrupted neutrophil function.11,15,19 Importantly, and in agreement with the data of Skokowa et al, in cells from patients suffering from SCN, characterized by complete lack of mature neutrophils, treatment with NAM increased the number of neutrophils. Here we demonstrate that this can be at least partially the result of increasing C/EBPε function (Figure 5B), thereby potentially compensating for decreased activity of the LEF1-C/EBPα axis in myeloid progenitors of patients with SCN.45 Together, the data presented in this study increase the knowledge of the functional regulation of C/EBPε activity and provide novel insights into mechanisms underlying aberrant neutrophil development. Modulating acetylation by NAM or other KDACi could therefore be of importance for the treatment of neutropenia or functional neutrophil defects.32,35,41
There is an Inside Blood Commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The work of M. Bartels was financially supported by a grant from the Wilhelmina Children's Hospital Research Fund. The work of A.M.G. and S.J.V. was funded by the Dutch Cancer Foundation, and V.F. and J.v.L. were financially supported by the Dutch Rheumatism Foundation.
Authorship
Contribution: M.B. designed the study, conducted the research, analyzed the data, and wrote the manuscript; A.M.G., V.F., R.v.G., and A.B.B. conducted the research and analyzed the data; A.R.L. and C.E.P. conducted the research; S.V. analyzed the data; M.B.B. reviewed the data; S.J.A. reviewed the data and edited the manuscript; J.v.L. designed the study, analyzed the data, and edited the manuscript; P.J.C. designed the study, analyzed the data, and wrote the manuscript; and all authors reviewed and approved the final draft.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Paul J. Coffer, Center for Molecular Medicine, Department of Cell Biology & Division of Pediatrics, University Medical Center, Heidelberglaan 100, 3584 CX Utrecht, The Netherlands; e-mail: p.j.coffer@umcutrecht.nl.

![Figure 5. Acetylation of C/EBPε K121 and K198 is functionally important during neutrophil differentiation. CD34+ cells were differentiated toward neutrophils in the absence or presence of 5 mM NAM. (A) In UCB-derived cells, neutrophil differentiation was determined by cytospin analysis of neutrophil morphology and fluorescence-activated cell sorter analysis of intracellular lactoferrin expression (mean fluorescence intensity [MFI]). (B) In SCN patient BM-derived cells, neutrophil development was determined by cytospin analysis of the percentage mature neutrophils and metamyelocytes. (C) At day 7 of differentiation, intracellular lactoferrin expression (MFI) was analyzed and protein lysates were prepared, followed by western blot analysis of C/EBPε expression. (D) CD34+ cells from UCB were retrovirally transduced with eGFP-PMX-C/EBPε, eGFP-PMX-C/EBPε eGFP-PMX-C/EBPε K121/198R, eGFP-PMX-C/EBPε K15xR, and C/EBPε R121/198K and differentiated toward neutrophils. As control, an empty vector was used. Sorted cells were analyzed after 14 days. Neutrophil differentiation was determined by cytospin analysis of morphology and fluorescence-activated cell sorter analysis of lactoferrin expression (MFI). (E) mRNA expression of lactoferrin and collagenase was analyzed by real-time quantitative polymerase chain reaction. Data represent the expression of lactoferrin and collagenase relative to the empty vector. Data are representative of 3 (or more) UCB donors or patient samples. Error bars represent standard error of the mean (between donors). *P < .05; **P < .01.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/11/10.1182_blood-2013-12-543850/4/m_1782f5.jpeg?Expires=1769108643&Signature=p9cg4bnmDLzBHLJKo-7Ezza2nE5GFfIT8VethaCg9~9sOesbYlMUUV-mntFSjlImK2JzHsIyFmLj25~y5RI-DOEAO-tX4AUjmAqrK2zmP9dLl~5xX~pTNySrmSmphShjyOn4Aa2bHYPtWAVMmX-~pES75AkcxzZHB~UfqXQqPSmh6zyxNSF4lN6fRN-BZ8y9E~v9Z2ubePjLJGR60eKL8hZUa4WsL-kLEG8auXuUr9B7NaTTEU3bYzpflXRBaYFVEX-ouTZHT9ZdFll94l~sxZ6dH2c8ZdnqdIg0HA~SOHVsEjtozpmwq~9AtNKSxJkauv7GgKQQOijdv3zmB0YwTA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal