Key Points
Benign (ie, IgM MGUS and smoldering WM) clonal B cells already harbor the phenotypic and molecular signatures of the malignant WM clone.
Multistep transformation from benign (ie, IgM MGUS and smoldering WM) to malignant WM may require specific copy number abnormalities.
Abstract
Although information about the molecular pathogenesis of Waldenström macroglobulinemia (WM) has significantly advanced, the precise cell of origin and the mechanisms behind WM transformation from immunoglobulin-M (IgM) monoclonal gammopathy of undetermined significance (MGUS) remain undetermined. Here, we undertook an integrative phenotypic, molecular, and genomic approach to study clonal B cells from newly diagnosed patients with IgM MGUS (n = 22), smoldering (n = 16), and symptomatic WM (n = 11). Through principal component analysis of multidimensional flow cytometry data, we demonstrated highly overlapping phenotypic profiles for clonal B cells from IgM MGUS, smoldering, and symptomatic WM patients. Similarly, virtually no genes were significantly deregulated between fluorescence-activated cell sorter-sorted clonal B cells from the 3 disease groups. Interestingly, the transcriptome of the Waldenström B-cell clone was highly different than that of normal CD25−CD22+ B cells, whereas significantly less genes were differentially expressed and specific WM pathways normalized once the transcriptome of the Waldenström B-cell clone was compared with its normal phenotypic (CD25+CD22+low) B-cell counterpart. The frequency of specific copy number abnormalities [+4, del(6q23.3-6q25.3), +12, and +18q11-18q23] progressively increased from IgM MGUS and smoldering WM vs symptomatic WM (18% vs 20% and 73%, respectively; P = .008), suggesting a multistep transformation of clonal B cells that, albeit benign (ie, IgM MGUS and smoldering WM), already harbor the phenotypic and molecular signatures of the malignant Waldenström clone.
Introduction
Waldenström macroglobulinemia (WM) is a lymphoplasmacytic lymphoma associated with a monoclonal immunoglobulin (Ig)M protein and bone marrow (BM) infiltration by small lymphocytes that may exhibit plasma cell differentiation.1 The presence vs absence of disease-associated symptoms defines symptomatic and smoldering WM,2 whereas those patients with an IgM paraprotein but no symptoms and no BM infiltration are diagnosed with an IgM monoclonal gammopathy of undetermined significance (MGUS).2 Because both smoldering WM and IgM MGUS patients are at risk of developing symptomatic WM,3,4 it could be speculated that similarly to multiple myeloma (MM),5 most (if not all) WM patients have eventually gone through the benign stages of IgM MGUS and smoldering WM before developing clinical symptoms.6
The pivotal study by Treon et al7 showing that MYD88 was recurrently mutated in WM patients (91%) but less frequently in IgM MGUS (10%) suggested that the MYD88 mutation could represent a critical event for progression of IgM MGUS to WM.8 However, although subsequent studies using sensitive allele specific oligonucleotide–polymerase chain reaction (ASO-PCR) confirmed the high prevalence of MYD88 L265P in WM (87-100%), they also unraveled that the MYD88 mutation is present in a much higher fraction (at least half) of IgM MGUS patients.9-12 These results indicate that, although the MYD88 mutation may be considered as a unifying event in the pathogenesis of WM, MYD88 by itself is insufficient to explain the malignant transformation of IgM MGUS to WM. Additional genetic abnormalities have been investigated in symptomatic WM,13-20 but in contrast to MM, there are virtually no data on the genomic landscape of clonal B cells from smoldering WM and IgM MGUS patients.13,19,21 Similarly, information about the transcriptome of the Waldenström clone is very limited and mostly restricted to symptomatic patients.22-24 Thus, the mechanism behind the malignant transformation of WM remains unknown.
Although conventional diagnosis of IgM MGUS requires the absence of BM infiltration in a trephine biopsy,2 the presence of MYD88 mutations indicates that a small B-cell clone may already be present. In line with this hypothesis, we recently observed the presence of clonal B cells in IgM MGUS (12% of patients) that progressively accumulate in smoldering and symptomatic WM.6 Such progressive accumulation of clonal B cells was accompanied by the emergence of a characteristic Waldenström phenotype (CD22+lowCD25+CD27+IgM+)6 ; however, because at the time we were limited by conventional 4-color flow cytometry, it was not possible to determine whether the emergence of a characteristic Waldenström phenotype from IgM MGUS to WM was due to progressive accumulation of clonal B cells or to the phenotypic transformation of clonal, yet benign, B cells, into full malignant tumor B cells.
Here, we used novel multidimensional (17-color) flow cytometry for sensitive detection and phenotypic characterization of clonal B cells in all stages of the disease, from IgM MGUS to smoldering and symptomatic WM. Subsequently, we performed sensitive fluorescence-activated cell sorter (FACS) sorting to investigate and compare the molecular signature and genomic landscape of clonal B cells in the benign (IgM MGUS and smoldering WM) vs malignant (symptomatic WM) stages of the disease. Our results show that IgM MGUS and WM patients share clonal B cells with similar phenotypic and molecular signatures, but specific genetic alterations that are commonly present in WM are less frequently detected in IgM MGUS. The overall comparison of the gene expression profiles of clonal B cells vs distinct subsets of normal B cells suggests that CD25+CD22+low activated B cells could represent the cellular origin of the Waldenström clone.
Patients and methods
A total of 51 newly diagnosed patients with an IgM monoclonal gammopathy—classified as IgM MGUS (n = 20), smoldering WM (n = 20), and symptomatic WM (n = 11)—are the focus of the present study. IgM MGUS patients were classified according to absent symptomatology, serum IgM monoclonal protein < 30 g/L, and BM lymphoplasmacytic infiltration <10%.3,4,25 The presence of >10% lymphoplasmacytic cells in the BM aspirate classified WM patients, and the distinction between smoldering and symptomatic WM was based on the absence vs presence of symptoms related to tumor infiltration (typically anemia, hepatosplenomegaly, hiperviscosity, and/or peripheral neuropathy).6 Samples were collected prior to therapy after informed consent was given, following recommendations and guidelines of the local ethics committee and the Declaration of Helsinki.
Multidimensional flow cytometry immunophenotyping
Approximately 400 μL of EDTA-anticoagulated BM-aspirated samples were immunophenotyped using an 8-color direct immunofluorescence stain-and-then-lyse technique, with 4 different combinations of monoclonal antibodies (mAbs) (Pacific Blue [PacB]/Pacific Orange [PacO]/fluorescein isothiocyanate [FITC]/phycoerythrin [PE]/peridinin chlorophyll protein-cyanin 5.5 [PerCP-Cy5.5]/PE-cyanin 7 [PE-Cy7]/allophycocyanin [APC]/APCH7): (1) CD38/CD45/surface IgM (sIgM)/CD27/CD79b/CD19/intracytoplasmic κ (cyIgκ)/intracytoplasmic λ (cyIgλ); (2) CD38/CD45/CD20/CD25/CD22/CD19/cyIgκ/cyIgλ; (3) CD38/CD45/CD103/CD305/CD11c/CD19/cyIgκ/cyIgλ; and (4) CD38/CD45/CD10/CD200/CD5/CD19/cyIgκ/cyIgλ. Data acquisition was performed for around 106 nucleated cells per tube in a FACSCantoII flow cytometer (Becton Dickinson Biosciences [BDB], San Jose, CA) using the FACSDiva 6.1 software (BDB). Monitoring of instrument performance was performed daily using the Cytometer Setup Tracking (BDB) and rainbow 8-peak beads (Spherotech, Lake Forest, IL) after laser stabilization, following the EuroFlow guidelines26 ; sample acquisition was systematically performed after longitudinal instrument stability was confirmed (supplemental Figure 1, available on the Blood Web site).
Generation of immunophenotypic protein expression profiles
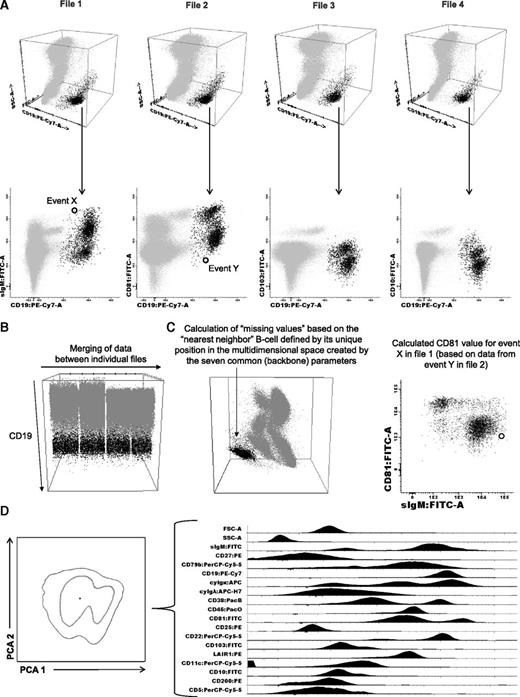
To generate immunophenotypic protein expression profiles (iPEPs), we first used those 7 parameters that were measured in common for each sample aliquot (CD38, CD45, CD19, cyIgκ, cyIgλ, forward light scatter [FSC], and sideward light scatter [SSC]) to define the B-cell compartment. Then, the merge function of the Infinicyt software (Cytognos SL, Salamanca, Spain) was used to fuse the different data files corresponding to the 4 different 8-color mAb combinations studied per sample, into a single data file containing all information measured for that sample (Figure 1A).27 Then, the calculation function of the Infinicyt software was used to fill in antigens that were not directly evaluated (“missing values”), based on the “nearest neighbor” statistical principle, defined by its unique position in the multidimensional space created by the 7 common (backbone) parameters (FSC/SSC/CD38/CD45/CD19/cyIgκ/cyIgλ; Figure 1B). Ultimately, an iPEP was generated for every single clonal B cell (Figure 1C), which included all 17 phenotypic markers analyzed plus FSC and SSC (Figure 1D).26 BM clonal B cells were systematically discriminated from normal B cells based on the presence of previously described aberrant phenotypes6,28 plus further confirmation of clonality through simultaneous assessment of light-chain restriction. The comparison between the iPEP of clonal B cells from the 3 disease stages was performed by principal component analysis (PCA), based on the 17 parameters evaluated under the automated population separator graphical representation of the Infinicyt software.26 Raw data used to generate the iPEP for each individual case are available in the supplemental Materials. The same approach was conducted to compare the iPEPs of patients with mutated vs wild-type MYD88 and the iPEP of clonal B cells from IgM MGUS plus WM cases vs clonal B cells from patients with marginal zone lymphoma (MZL; n = 8), and chronic lymphocytic leukemia (B-CLL; n = 7).
Generation of iPEPs. After identification of the B-cell compartment (A) within individual files, (B) merging of data allows for (C) calculation of phenotypic information contained in 4 different 8-color monoclonal antibody combinations (D) to create an iPEP that include 17 different markers plus FSC and SSC simultaneously analyzed on every single B cell.
Generation of iPEPs. After identification of the B-cell compartment (A) within individual files, (B) merging of data allows for (C) calculation of phenotypic information contained in 4 different 8-color monoclonal antibody combinations (D) to create an iPEP that include 17 different markers plus FSC and SSC simultaneously analyzed on every single B cell.
MYD88 L265P detection
DNA was analyzed for the presence of the MYD88 mutation in 26 of 51 cases (7 IgM MGUS, 14 smoldering, and 5 symptomatic WM) using a real-time allele-specific oligonucleotide PCR (ASO-RQ-PCR) to detect the MYD88 L265P mutation, following a previously validated and reported method.29
Copy number and gene expression analyses on FACS-sorted clonal B cells
Clonal B cells from IgM MGUS (n = 11), smoldering (n = 15), and symptomatic WM (n = 11) patients were FACS-sorted (FACSAria II, BDB; purity ≥97%) according to patient-specific aberrant phenotypes. Afterward, genome-wide detection of copy number abnormalities (CNAs) and loss of heterozygosity (LOH) were investigated using the standard Affymetrix Cytoscan 750K platform (Affymetrix, Santa Clara, CA). Given that matched normal DNA was only available in a subset of cases (n = 21), an unpaired analysis was performed using 240 Hapmap files. In those cases with paired normal DNA (peripheral blood T cells), the latter was also used to eliminate patient-specific CNAs. The complete data set was analyzed by visual inspection using the AGCC and ChAS software programs (Affymetrix). CNAs were reported when the 3e following criteria were met: ≥25 consecutive imbalanced markers per segment; ≥100-Kb minimum genomic size; and <50% overlap with paired control DNA and/or genomic variants of Toronto DB (DGV).30 Only copy number-neutral LOHs (CNN-LOHs) >5 Mb were considered.
Gene expression profiling (GEP) was performed in 4 IgM MGUS, 4 smoldering, and 6 symptomatic WM patients with adequate RNA extracted from FACS-purified clonal B cells. Additionally, CD25−CD22+ (n = 9) and CD22+lowCD25+ (n = 8) FACS-sorted normal BM B cells from 10 healthy donors were analyzed as controls. Briefly, the integrity of the extracted RNA was assessed using the Agilent 2100 Bioanalyzer. Afterward, RNA was amplified, labeled, and subsequently hybridized to the Human Gene 1.0 ST Array (Affymetrix).31 Normalization was carried out by using the expression console (Affymetrix) with the Robust Multi-array Average algorithm, which includes background correction, normalization, and calculation of expression values (log2).22 The SIMFIT (http://www.simfit.org.uk/) statistical software was used to perform hierarchical clustering analyses based on Euclidean distance as distances and the group average linkage method. Differentially expressed genes between classes were identified using the significant analysis of microarrays algorithm (http://statweb.stanford.edu/∼tibs/SAM/), and significant genes were selected based on the lowest q-value (<10−5).22,31 To identify the most relevant biological mechanisms, pathways, and functional categories involved according to differently expressed genes, the ingenuity pathway analysis (Ingenuity Systems, www.ingenuity.com) was used. Full microarray data are available at the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/; accession number GSE61597).
Statistical analysis
The Mann-Whitney U and χ2 tests were used to estimate the statistical significance of differences observed between groups, using SPSS software (version 15.0; SPSS, Chicago, IL).
Results
iPEP of the Waldenström clone
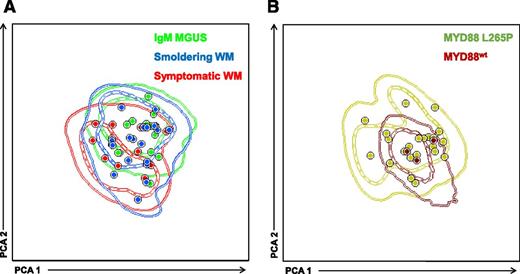
Through the sensitive multidimensional flow cytometry (MFC) approach described above (Figure 1), clonal B cells were detected in 15 of 20 (75%) IgM MGUS patients, all but 1 smoldering WM cases (19/20; 95%), and in all (100%) patients with symptomatic WM. Clonal B cells systematically showed sIgM+ expression, with a CD22+low and CD25+ phenotype homogeneously expressed in 81% and 89% of patients, respectively. Heterogeneous bimodal (±) patterns of expression were observed for CD27 (51% of patients), CD38 (50%), and CD200 (62%). CD79b and CD81 were positive in all cases (though dim staining was detected in 5% and 19% of cases, respectively), whereas CD5, CD10, CD11c, and CD103 were mostly negative (95%, 100%, 96%, and 100% of patients, respectively). CD305 (LAIR1) was particularly useful to detect light-chain restricted clonal B cells due to its homogenous lack of expression in 69% of cases, which contrasts to the bimodal heterogeneous staining on normal B cells. No significant differences were observed in the pattern of antigen expression (ie, negative, bimodal, and/or positive), as well as the percentage of positive cells for each individual marker between clonal B cells from IgM MGUS, smoldering, and symptomatic WM patients. Accordingly, PCA of merged iPEPs from each patient clonal B cells showed a complete phenotypic overlap between IgM MGUS, smoldering, and symptomatic WM patients (Figure 2A). Collectively, these results show that clonal B cells are phenotypically very similar among the 3 disease stages and that, on phenotypic grounds, the Waldenström clone is already detectable in IgM MGUS patients.
iPEP of clonal B cells from IgM MGUS, smoldering WM, and symptomatic WM according to disease stage and MYD88 mutational status. (A) iPEP of clonal B cells from IgM MGUS (n = 15; green), smoldering WM (n = 19; blue), and symptomatic WM (n = 11; red) patients. In the PCA graphic view, every patient is represented by a single dot and disease reference groups by 1 (dash lines) and 2 (solid lines) standard deviation curves. (B) PCA of iPEP of clonal B cells from patients with MYD88 mutated (n = 22; dark green dots and lines) vs wild-type MYD88 (n = 3; brown dots and lines).
iPEP of clonal B cells from IgM MGUS, smoldering WM, and symptomatic WM according to disease stage and MYD88 mutational status. (A) iPEP of clonal B cells from IgM MGUS (n = 15; green), smoldering WM (n = 19; blue), and symptomatic WM (n = 11; red) patients. In the PCA graphic view, every patient is represented by a single dot and disease reference groups by 1 (dash lines) and 2 (solid lines) standard deviation curves. (B) PCA of iPEP of clonal B cells from patients with MYD88 mutated (n = 22; dark green dots and lines) vs wild-type MYD88 (n = 3; brown dots and lines).
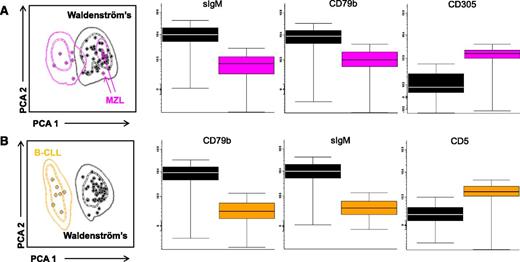
After demonstrating a common iPEP on clonal B cells from IgM MGUS and WM patients, we sought to investigate whether clonality could still be detected in patients with wild-type MYD88 and, in such case, to compare the iPEP of clonal B cells from patients with mutated vs wild-type MYD88. Clonality was detected in all cases with MYD88 L265P and also in 3 of 4 patients with nonmutated MYD88. Although the numbers are small to reach definitive conclusions, PCA comparing the iPEP of the 3 patients with wild-type MYD88 vs cases with mutated MYD88 showed fully overlapping phenotypes (Figure 2B). Overall, these results suggest that the unique phenotypic characteristics of Waldenström clonal B cells could potentially be of help on the differential diagnosis between WM and other B cell lymphoproliferative disorders, even in the absence of mutated MYD88. To demonstrate this, we compared the iPEPs of all patients with IgM MGUS, smoldering, and symptomatic WM patients (hereafter represented as the Waldenström clone) vs clonal B cells from lymphoproliferative disorders in which the MYD88 mutation can be detected (eg, MZL and B-CLL). PCA comparison between the Waldenström clone and tumor cells from patients with MZL showed divergent iPEPs in 6 of the 8 MZL cases (outside 2 standard deviations of the Waldenström iPEP), whereas the remaining 2 MZL patients had overlapping iPEP with that of Waldenström (Figure 3A). Interestingly, the 2 latter patients had nodal and extranodal mucosa-associated lymphoid tissue: MALT-MZL subtypes. The most significant (P < .05) markers to discriminate the Waldenström clone vs MZL were sIgM, CD79b, and CD305, the first 2 markers being overexpressed in IgM MGUS/WM, whereas CD305 was upregulated in MZL clonal B cells (Figure 3A). PCA comparison between IgM MGUS/WM and B-CLL showed completely separated iPEPs (Figure 3B), with the most significant markers being sIgM, CD79b (both upregulated in Waldenström clonal B cells), and CD5 (overexpressed in B-CLL).
PCA-based classification model for the differential diagnosis between the Waldenström clone (pooled data from IgM MGUS, smoldering, and symptomatic WM patients) vs clonal B cells from patients with MZL and B-CLL. The mean intensity fluorescence of the top 3 markers to discriminate between iPEPs from Waldenström vs clonal B cells from (A) MZL and (B) CLL patients are illustrated by the respective box plots. The pink arrows in A denote MZL patients inside standard deviation curves of the Waldenström clone iPEP.
PCA-based classification model for the differential diagnosis between the Waldenström clone (pooled data from IgM MGUS, smoldering, and symptomatic WM patients) vs clonal B cells from patients with MZL and B-CLL. The mean intensity fluorescence of the top 3 markers to discriminate between iPEPs from Waldenström vs clonal B cells from (A) MZL and (B) CLL patients are illustrated by the respective box plots. The pink arrows in A denote MZL patients inside standard deviation curves of the Waldenström clone iPEP.
GEP of the Waldenström clone vs distinct normal B-cell subsets
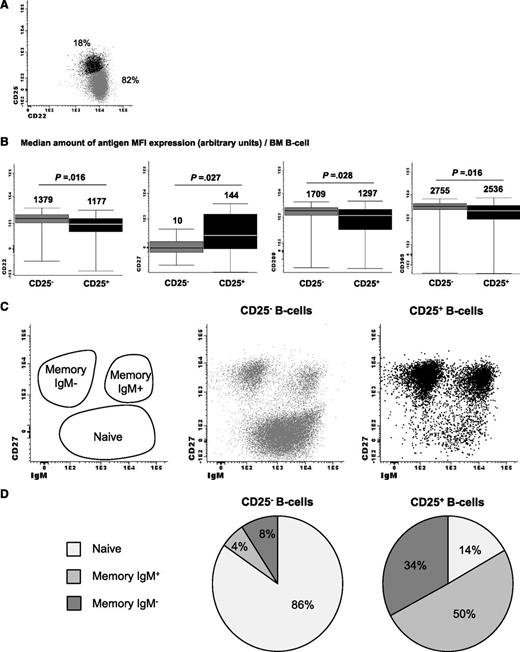
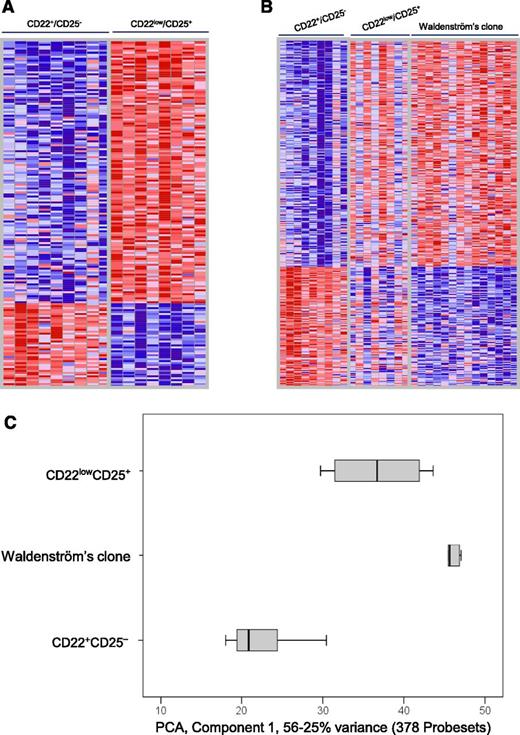
Because virtually no differences were observed between the GEP of clonal B cells from IgM MGUS and WM patients (data not shown), we decided to pool them together (hereafter referred as the Waldenström clone) and compare their GEP against that of normal B cells. Given the unique Waldenström phenotype described above, this comparison was performed against 2 well-defined subsets of normal BM mature B-lymphocytes: CD25− and CD25+ B cells (Figure 4A). Noteworthy, the phenotype of CD25+ normal B cells slightly differed from that of the CD25− subset, with significantly higher levels of CD27 associated with lower staining for CD22, CD200, and CD305 (P < .05; Figure 4B). Moreover, CD25+ B cells were significantly enriched on IgM memory B cells (P = .008; Figure 4C-D) compared with CD25− B cells. Interestingly, up to 177 genes were found to be differentially expressed genes between CD22+CD25− and CD22lowCD25+ normal BM B cells (Figure 5A). As expected, the highest upregulated gene on CD22lowCD25+ B cells was IL2RA (CD25), but also CD27 was found to be overexpressed, whereas CD200 was downregulated compared with CD22+CD25− cells. Accordingly, there was an accurate correlation between the results obtained at the protein and RNA levels for antigenic markers. Several genes (up to 32) implicated in B-cell activation/differentiation or B-cell lymphomagenesis were also found to be deregulated (supplemental Table 1). Interestingly, almost half of the genes differentially expressed between CD22lowCD25+ vs CD22+CD25− B cells (77/177, 44%) were also deregulated when the Waldenström clone was specifically compared with the CD22+CD25− B-cell subset (supplemental Table 1).
Phenotypic characterization of CD2+CD25− normal vs CD22lowCD25+ activated bone marrow B lymphocytes. Flow cytometry analysis of (A) CD22+CD25− vs CD22lowCD25+ normal bone marrow B lymphocytes and (B) paired comparison of mean fluorescence intensity of CD22, CD27, CD200, and CD305 expression, as well as (C-D) their corresponding distribution within the naïve, IgM+, and IgM− memory B-cell compartments.
Phenotypic characterization of CD2+CD25− normal vs CD22lowCD25+ activated bone marrow B lymphocytes. Flow cytometry analysis of (A) CD22+CD25− vs CD22lowCD25+ normal bone marrow B lymphocytes and (B) paired comparison of mean fluorescence intensity of CD22, CD27, CD200, and CD305 expression, as well as (C-D) their corresponding distribution within the naïve, IgM+, and IgM− memory B-cell compartments.
GEP of the Waldenström clone vs distinct normal B-cell subsets. (A) Heat map of genes with differential expression (false discovery rate q-value <10−5) between CD22+CD25− (n = 9) and CD22lowCD25+ (n = 8) normal bone marrow B-cell subsets. (B) Heat map of 634 genes deregulated (q-value <10−5) in Waldenström vs CD22+CD25− normal B cells and corresponding expression values on CD22lowCD25+. (C) PCA based on the sum of nonoverlapping genes (n = 799) differentially expressed between Waldenström and CD22+CD25− normal B cells plus genes differentially expressed between Waldenström and CD22lowCD25+ activated B cells, displayed (along the first principal component) by notched boxes representing 25th and 75th percentile values; the line in the middle and vertical lines correspond to the median value and both the 10th and 90th percentiles, respectively. The color bar depicts normalized intensity values ranging from low (dark blue) to high (dark red) expression levels.
GEP of the Waldenström clone vs distinct normal B-cell subsets. (A) Heat map of genes with differential expression (false discovery rate q-value <10−5) between CD22+CD25− (n = 9) and CD22lowCD25+ (n = 8) normal bone marrow B-cell subsets. (B) Heat map of 634 genes deregulated (q-value <10−5) in Waldenström vs CD22+CD25− normal B cells and corresponding expression values on CD22lowCD25+. (C) PCA based on the sum of nonoverlapping genes (n = 799) differentially expressed between Waldenström and CD22+CD25− normal B cells plus genes differentially expressed between Waldenström and CD22lowCD25+ activated B cells, displayed (along the first principal component) by notched boxes representing 25th and 75th percentile values; the line in the middle and vertical lines correspond to the median value and both the 10th and 90th percentiles, respectively. The color bar depicts normalized intensity values ranging from low (dark blue) to high (dark red) expression levels.
A total of 327 genes were differentially expressed between the Waldenström clone and normal CD22+CD25− B cells (Figure 5B; supplemental Table 2). In addition to disease-related genes (eg, BACH2, BCLAF1, IL4R, IL21R, TRAF3, TNFRS10B), IL2RA (CD25) and CD27 were found to be overexpressed, whereas CD200 and LAIR1 (CD305) downregulated in Waldenström vs CD22+CD25− B cells, confirming once more the correlation between iPEP and GEP analyses. By contrast, differences in GEP were much less evident on comparing the Waldenström clone vs CD22lowCD25+ normal B cells, with only 51 genes (instead of the previous 327) being downregulated (and none overexpressed) in Waldenström cells (supplemental Table 3). Interestingly, enrichment analysis unraveled that specific biofunctions, pathways, and upstream regulators were similarly altered while comparing CD22lowCD25+ vs CD22+CD25− normal B cells and Waldenström vs CD22+CD25− B lymphocytes, but not on comparing the Waldenström clone vs CD22lowCD25+ (Table 1). These results suggest ongoing activation of WM-related pathways in normal activated (CD22lowCD25+) B lymphocytes, and in fact, some biofunctions or upstream regulators were found to be more active in CD22lowCD25+ B cells vs the Waldenström clone. Altogether, these results suggest that CD22lowCD25+ (activated) B cells are genetically closer to WM and could represent the physiological counterpart of WM (Figure 5C).
Selected biofunctions, canonical pathways, and upstream regulators predicted to be significantly activated or inhibited according to the corresponding deregulated genes between CD22+CD25− and CD22lowCD25+ normal bone marrow B lymphocytes and between the Waldenström clone vs each normal B-cell subset
| Enrichment analysis . | CD22lowCD25+ vs CD22+CD25− . | Waldenström clone vs CD22+CD25− B cells . | Waldenström clone vs CD22lowCD25+ B cells . |
|---|---|---|---|
| Biofunctions | |||
| Cellular development, cellular growth and proliferation | Activated (26 genes; P = 9.7E-07) | Activated (39 genes; P = 2.8E-07) | Inhibited (18 genes; P = 3.6E-02) |
| Cell death and survival | Activated (57 genes; P = 3.7E-04) | Activated (46 genes; P = 2.7E-04) | — |
| Humoral immune response, IgG protein synthesis | Inhibited (9 genes; P = 2.0E-04) | Inhibited (13 genes; P = 7.2E-05) | — |
| Canonical pathways | |||
| D-myo-inositol (1,3,4,5,6)-tetrakisphosphate biosynthesis | Activated (6 genes; P = 6.4E-04) | Activated (8 genes; P = 5.8E-04) | — |
| 3-phosphoinositide biosynthesis | Activated (7 genes; P = 3.3E-04) | Activated (8 genes; P = 2.4E-03) | — |
| Upstream regulators | |||
| BCR | Activated (8 genes; P = 1.4E-06) | Activated (12 genes; P = 2.6E-08) | — |
| CD40 | Activated (10 genes; P = 1.2E-06) | Activated (13 genes; P = 1.5E-06) | — |
| INFα | Activated (12 genes; P = 5.8E-06) | Activated (14 genes; P = 1.6E-04) | — |
| INFγ | Activated (19 genes; P = 2.2E-03) | Activated (40 genes; P = 3.8E-07) | Inhibited (9 genes; P = 1.2E-03) |
| miRNA-92a | Inhibited (20 genes; P = 9.3E-04) | Inhibited (38 genes; P = 2.8E-06) | — |
| miRNA-181a | Inhibited (24 genes; P = 9.8 E-04) | Inhibited (49 genes; P = 1.3E-07) | — |
| TP53 | Activated (23 genes; P = 3.7E-04) | Activated (36 genes; P = 6.0E-05) | — |
| Enrichment analysis . | CD22lowCD25+ vs CD22+CD25− . | Waldenström clone vs CD22+CD25− B cells . | Waldenström clone vs CD22lowCD25+ B cells . |
|---|---|---|---|
| Biofunctions | |||
| Cellular development, cellular growth and proliferation | Activated (26 genes; P = 9.7E-07) | Activated (39 genes; P = 2.8E-07) | Inhibited (18 genes; P = 3.6E-02) |
| Cell death and survival | Activated (57 genes; P = 3.7E-04) | Activated (46 genes; P = 2.7E-04) | — |
| Humoral immune response, IgG protein synthesis | Inhibited (9 genes; P = 2.0E-04) | Inhibited (13 genes; P = 7.2E-05) | — |
| Canonical pathways | |||
| D-myo-inositol (1,3,4,5,6)-tetrakisphosphate biosynthesis | Activated (6 genes; P = 6.4E-04) | Activated (8 genes; P = 5.8E-04) | — |
| 3-phosphoinositide biosynthesis | Activated (7 genes; P = 3.3E-04) | Activated (8 genes; P = 2.4E-03) | — |
| Upstream regulators | |||
| BCR | Activated (8 genes; P = 1.4E-06) | Activated (12 genes; P = 2.6E-08) | — |
| CD40 | Activated (10 genes; P = 1.2E-06) | Activated (13 genes; P = 1.5E-06) | — |
| INFα | Activated (12 genes; P = 5.8E-06) | Activated (14 genes; P = 1.6E-04) | — |
| INFγ | Activated (19 genes; P = 2.2E-03) | Activated (40 genes; P = 3.8E-07) | Inhibited (9 genes; P = 1.2E-03) |
| miRNA-92a | Inhibited (20 genes; P = 9.3E-04) | Inhibited (38 genes; P = 2.8E-06) | — |
| miRNA-181a | Inhibited (24 genes; P = 9.8 E-04) | Inhibited (49 genes; P = 1.3E-07) | — |
| TP53 | Activated (23 genes; P = 3.7E-04) | Activated (36 genes; P = 6.0E-05) | — |
A detailed list of genes under each selected biofunction, canonical pathway, and upstream regulator and their corresponding expression status is available in supplemental Table 5.
Genomic landscape of the Waldenström clone
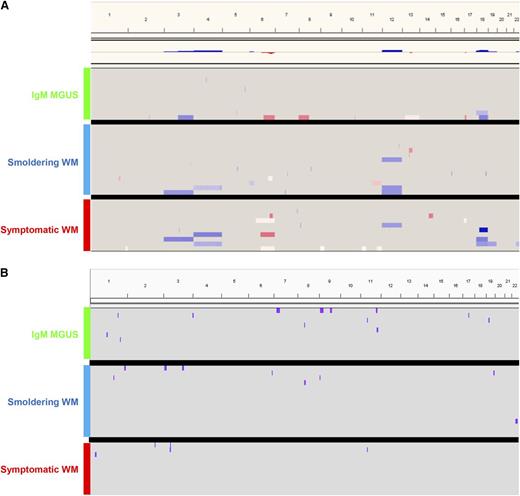
Overall, 24 of 37 patients (65%) displayed chromosomal abnormalities. A total of 53 CNAs were detected (32 gains and 21 losses) in FACS-purified clonal B cells from IgM MGUS and WM patients, with a median of 2 imbalances per altered case (range, 1-9). The frequency of patients displaying CNAs significantly increased (P = .03) from IgM MGUS (4/11; 36%) to smoldering (11/15; 73%) and symptomatic WM (9/11; 82%; Figure 6A); by contrast, the frequency of CNN-LOH did not significantly differ according to disease stage (Figure 6B). All chromosomal imbalances, gains and deletions of chromosomal arms, and interstitial CNAs per disease stage are detailed in supplemental Table 4, and a list of genes under all CNN-LOHs identified is provided in supplemental Materials. The 2 most common minimally altered regions (≥10% of patients) present in all disease stages consisted of del(6q23.3-6q25.3) and +18q22.1, accounting for 16% and 14% of patients, respectively. However, differences between the 3 disease stages were also observed; thus, genomic imbalances typically observed in WM (6q− or 18q+) were rarely seen in clonal B cells from IgM MGUS patients. Moreover, trisomy 4 and trisomy 12 were exclusively detected in smoldering and symptomatic WM, whereas they were absent in IgM MGUS cases (Figure 6A). Accordingly, up to 73% of symptomatic WM had ≥1 of the most common CNAs, +4, del(6q23.3-6q25.3), +12, and +18q22.1, whereas their frequency in IgM MGUS and smoldering WM cases was significantly inferior (18% and 20%, respectively; P = .008). A detailed list of genes under the 4 most common CNAs can be found in supplemental Materials. Collectively, our results indicate that the number of CNAs in WM is rather low but also that specific genetic lesions involving chromosomes 4 and 12, as well as on the 6q23.3-6q25.3 and +18q21.1 regions, could be associated with malignant transformation of the Waldenström clone.
Genomic landscape of the Waldenström clone. Overview of (A) CNAs and (B) CNN-LOH detected in clonal B cells from IgM MGUS (n = 11), smoldering WM (n = 15), and symptomatic WM (n = 11) patients.
Genomic landscape of the Waldenström clone. Overview of (A) CNAs and (B) CNN-LOH detected in clonal B cells from IgM MGUS (n = 11), smoldering WM (n = 15), and symptomatic WM (n = 11) patients.
Discussion
In contrast to MM, where significant efforts have been done to characterize the premalignant stages of the disease (ie, MGUS and smoldering MM),31-35 no high-throughput studies have been performed on clonal B cells from IgM MGUS, smoldering, and symptomatic WM patients to understand the mechanisms behind disease progression. Here, we undertook an integrated phenotypic, molecular, and genomic approach to study clonal B cells from each disease stage. Our results denote a common phenotypic and molecular signature for IgM MGUS and WM (albeit with less genetic abnormalities in the early stages), suggesting a common cellular origin (ie, CD25+CD22+low activated B lymphocytes) and a potential unifying genetic event (eg, MYD88 L265P) rendering the WM signature.
After the initial findings by Treon et al that resulted in the identification of a recurrent somatic mutation (L265P) in WM involving the MYD88 gene,7 additional studies have confirmed and extended such observations to IgM MGUS (with 50-80% of mutated cases),9,10,12 thereby confirming its role as an early oncogenic event. Among other effects, MYD88 activating mutations promote nuclear factor-κB signaling, Janus kinase (JAK) activation of signal transducer and activator of transcription 3 (STAT3), and secretion of interleukin-6.36 Such mutation is likely to induce a disease-specific transcriptional signature; in fact, 2 independent studies have reported that the GEP of WM patients is markedly homogeneous.22,23 Our results, based on the specific comparison between clonal B cells from IgM MGUS and WM patients vs normal resting (CD25−CD22+) B cells showed deregulated genes related to the IL-6 (eg, ABCB1, GRB2), NF-κB (eg, TRAF3, IGF1R, BMPR2, TBK1), JAK/STAT (eg, GRB2, PTPN1), and phosphatidylinositol 3-kinase/protein kinase B (PI3K/AKT) (eg, GAB2, GRB2, CDKN1B, IL4R, ITPR2, ITPR1) signaling pathways, which have been previously implicated in the growth and survival of WM cells.16,22,23,37-41 Moreover, we unravel additional deregulated pathways such as inositol tetrakisphosphate and 3-phosphoinositide biosynthesis (Table 1). Such altered pathways would support the notion of WM arising from an antigen-experienced recently activated B cell that exits the germinal center42 and cannot fully mature into terminally differentiated plasma cells.43,44
The knowledge about the molecular pathogenesis of WM has greatly improved in recent years,21 but the exact cellular origin of the Waldenström clone remains to be determined.39,44 The presence of somatic hypermutations would suggest a memory B cell of origin; however, absence of CD27 and somatic hypermutations in a subgroup of WM patients6,39,45,46 would favor, at least in the latter cases, a nongerminal center cellular origin. To shed some light into this open question, we compared the molecular profile of Waldenström cells vs their normal phenotypic counterpart: CD22lowCD25+ B cells. In normal activated B lymphocytes, expression of CD25 rapidly increases following Toll-like receptor (TLR)-mediated cell stimulation, resulting in the expression of a fully functional IL-2R; this mechanism of IL-2R activation is NF-κB dependent.47 Interestingly, many of the genes deregulated between Waldenström and CD22+CD25− normal B cells were expressed at similar levels on CD22lowCD25+ B lymphocytes; in fact, the number of genes differentially expressed between the Waldenström clone and CD22lowCD25+ B cells was dramatically reduced compared with the analysis against the CD22+CD25− subset. Furthermore, individual comparison between the Waldenström clone and each B-cell subset unraveled that CD22lowCD25+ B cells showed similar activation status of upstream regulators such as the B-cell receptor CD40, miRNA-92a, and miRNA-181a (both recently implicated in TLR regulation48,49 ), and even several genes related to cellular development, growth and proliferation, as well as genes downstream of the interferon γ regulator were found to be more active in CD22lowCD25+ B-cells compared to the Waldenström clone. Interferon γ-producing B cells have been recently described as a new subset that can promote innate responses against intracellular infection, via increased Bruton tyrosine kinase signaling and NF-κB activation, after CD40 ligation.50 Of note, 4 large nationwide studies exploring the role of antigenic stimulation in the pathogenesis of WM have shown a notably higher risk among individuals with infectious diseases.51-54 Altogether, these results support further functional analyses exploring the role of chronic antigenic stimulation in the etiology of WM.
One of the major aims of the present study was to compare the molecular and phenotypic features of clonal B cells from IgM MGUS vs WM patients for the potential identification of such events related with malignant transformation of the disease. Although the number of patients precludes definitive conclusions, no major differences were observed between gene expression levels of clonal B cells from the 3 disease stages (data not shown), in line with preliminary findings by Chng et al.23 Interestingly, however, the expression levels of the AT-rich interactive domain 5B (ARID5B) gene was fivefold higher in smoldering WM vs IgM MGUS; furthermore, ARID3B was also found to be overexpressed in Waldenström vs CD22+CD25− normal B cells. Hunter et al55 recently found that ARID1A was the third most common target gene for mutations in WM, and loss of the ARID1B alternate family member was the most frequent target of del(6q).55 Thus, further investigations are warranted to define the exact role of this class of molecules in the pathogenesis of WM. In line with the GEP analyses, the iPEP of clonal B cells from IgM MGUS, smoldering, and symptomatic WM was also highly similar. These findings confirm our recent observations6 and establish a WM-specific phenotype, which could be particularly useful for the differential diagnosis of MYD88 wild-type WM patients, as well as of other B-cell disorders with mutated MYD88 (ie, MZL). Accordingly, clonal B cells were still detected by MFC among cases with nonmutated MYD88, and, albeit the low numbers, these preliminary results suggest that their iPEP is superimposable to that of MYD88 L265P patients. Noteworthy, by using a sensitive MFC method, clonal B cells were detected in 75% of cases, in line with the results obtained by sensitive molecular screening of MYD88 L265P in IgM MGUS patients.9 Altogether, these results support diagnostic criteria based on the extent of BM infiltration3,4,25,56,57 rather than only the presence vs absence of clonality on trephine biopsies for differential diagnosis between IgM MGUS and WM.
If the MYD88 mutation is indeed an early oncogenic event,55 then additional genetic hits are likely to be required for the malignant transformation of WM. To the best of our knowledge, only in 1 study have chromosomal abnormalities been investigated among smoldering WM patients,58 and no data are available on IgM MGUS cases. Here, we compared the genomic profile of FACS-sorted clonal B cells from the 3 disease stages; our results showed a significantly increased frequency of patients displaying genetic abnormalities in the transition from IgM MGUS to symptomatic WM. However, in line with previous observations,16,18,58 we confirmed that the genomic landscape of WM is less complex than that of MM (median of 2 vs 12 CNAs per altered case, respectively).35 It could well be that rather than an increasing number of CNAs, specific genetic abnormalities [eg, +4, del(6q23.3-6q25.3), +12, and +18q22.1] are required for the malignant transformation of WM. Interestingly, recent whole-genome sequencing studies have demonstrated that small CNAs involving B-cell regulatory genes are highly prevalent in WM,55 and many of those genes are coded within the most frequently altered chromosomal regions in the present study (eg, HIVEP2, PLEKHG1, ARID1B located in the 6q23.3-6q25.3 deleted region). These findings suggest a multistep process for WM evolution from IgM MGUS, wherein a common genetic event (ie, the MYD88 mutation) occurs in the B-cell subset with a highly specific (activated) phenotype (ie, CD25+CD22+low) and enriched on IgM+ memory B cells (Figures 5A and 6), that would help the clone to avoid apoptosis59 (in fact, genes downstream of TP53 were commonly expressed in Waldenström and CD22lowCD25+ cells) and prolong the upregulation of cellular growth and proliferation genes; additional genetic abnormalities would then be potentially required for this small B-cell clone to expand and produce higher levels of IgM and for the patient to develop clinical symptoms. Accordingly, deletion of 6q, the progressive expansion and proportion of clonal over normal B cells, as well as increased IgM levels, are prognostic factors to predict risk of transformation of IgM MGUS and smoldering WM into symptomatic WM.3,6,15,60
In summary, our results show that IgM MGUS patients have clonal B cells that, albeit less CNAs, are phenotypically and molecularly similar to those of WM patients. The overall gene expression profile of the Waldenström clone vs CD25−CD22+ resting B lymphocytes and CD25+CD22+low activated B cells suggest the latter as the cellular origin of the disease.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by Cooperative Research Thematic Network grants RD12/0036/0058 and RD12/0036/0048 of the Red de Cancer (Cancer Network of Excellence), Consejerı´a de Sanidad, Junta de Castilla y Leon, Valladolid, Spain (557/A/10).
Authorship
Contribution: J.F.S.M. and B.P. conceived the idea and designed the study; B.P., M.-B.V., J.J.P., and N.P. analyzed flow cytometric data; I.A.-M. performed flow cytometry immunophenotyping; M.-L.S., P.B., and D.A. performed cell sorting; C.J. and M.-E.S. performed ASO-PCR studies; M.-V.M., E.M.O., F.E., J.H., R.C., A.G.d.C., M.S., M.-C.M., T.J.G.-L., J.G., A.B., J.A., E.P., N.C.G., and J.F.S.M. supplied study material or patients; B.P. and L.A.C. performed statistical analysis; B.P., A.O., and J.F.S.M. analyzed and interpreted data; B.P., A.O., and J.F.S.M. wrote the manuscript; and all authors reviewed and approved the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Bruno Paiva, Clinica Universidad de Navarra, Centro de Investigacion Médica Aplicada (CIMA), Av. Pio XII 36, 31008 Pamplona, Spain; e-mail: bpaiva@unav.es.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal