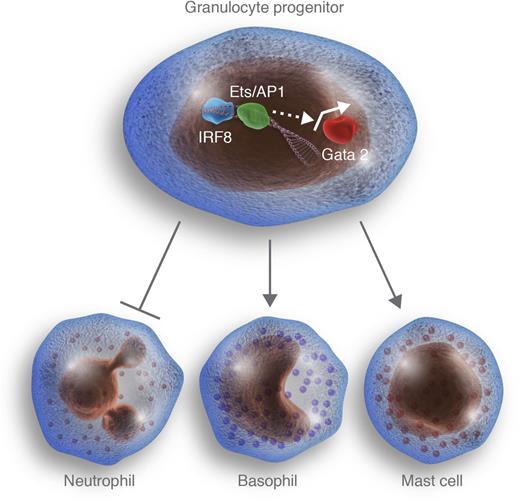
In this issue of Blood, Sasaki and colleagues demonstrate a strict requirement for the transcription factor interferon regulator factor 8 (IRF8) in the development of mouse basophils.1
IRF8 represses neutrophil potential and promotes basophil and mast cell development. Sasaki and colleagues demonstrate that the transcription factor IRF8 interacts with transcriptional partners, such as Ets and/or AP-1 family members, to activate Gata2 expression in granulocytic progenitors, probably through indirect mechanisms. The expression of Gata2 in turn represses production of neutrophils and promotes basophil and mast cell production. Professional illustration by Luk Cox, Somersault 18:24.
IRF8 represses neutrophil potential and promotes basophil and mast cell development. Sasaki and colleagues demonstrate that the transcription factor IRF8 interacts with transcriptional partners, such as Ets and/or AP-1 family members, to activate Gata2 expression in granulocytic progenitors, probably through indirect mechanisms. The expression of Gata2 in turn represses production of neutrophils and promotes basophil and mast cell production. Professional illustration by Luk Cox, Somersault 18:24.
Basophils represent the rarest type of circulating granulocyte, comprising approximately 1% of blood leukocytes. Basophils are present and well conserved across species, strongly suggesting important functional roles. As robust markers and mouse models have become available, it is now clear that basophils play important roles in both Th2-mediated immunity to pathogenic helminths and atopic inflammation,2 yet the cellular steps and molecular requirements for the development of basophils are not fully understood.
Delineating cellular pathways in hematopoiesis traditionally involves the prospective isolation of progenitors using panels of cell-surface markers. These progenitors can then be assessed functionally through in vivo transplantation, clonal assays in vitro, and gene expression analysis. From a translational perspective, committed proximal progenitors can give rise to functional mature cells much more rapidly than do hematopoietic stem cells after transplantation. This in turn can provide rapid immunity to specific types of infections following immune suppression. Given evidence that they can readily cross allogeneic barriers under these conditions, the clinical potential of using progenitors as off-the-shelf cellular therapies is tantalizing.3
Basophil progenitors are likely derived from myeloid precursors with pan-granulocytic and monocytic developmental potential, yet the precise subsequent cellular stages that lead to a fully committed basophil are not fully resolved. Specifically, it remains unclear whether a bipotent basophil/mast cell progenitor exists or whether the mast cell lineage diverges prior to the restriction of other granulocytic lineages.4,5 At the heart of this controversy is the assumption that commitment to a lineage must proceed through a single cellular route and that progenitors with partially overlapping potentials cannot coexist. For example, would the existence of a bipotent basophil/mast cell–committed progenitor obligatorily preclude the existence of a bipotent basophil/neutrophil progenitor? In all likelihood, this cannot be assumed. Indeed, the transcription factor IRF8 can generate progenitors with partially overlapping potentials by enacting developmental programs autonomously of cellular context.6 Thus, the application of genetic approaches to hematopoietic progenitor studies can help resolve controversies in cellular developmental models.
Sasaki and colleagues now demonstrate that IRF8 is essential for the development of mouse basophils.1 Using a knockin green fluorescent protein reporter, the authors observe that IRF8 expression cannot be detected in mature basophils and is instead found within granulocyte-macrophage progenitors (GMPs). The authors find that Irf8−/− GMPs overproduce neutrophils but fail to generate basophils. Epistatic experiments show that IRF8 acts in part by increasing the expression of another transcription factor, GATA2. Interestingly, the authors find that mast cell potential is also adversely affected by IRF8 deficiency. These data suggest, but certainly do not prove, the existence of a common genetic program and/or progenitor between the basophil and mast cell lineages.
IRF8 plays important roles in a number of other fate decisions within the myeloid compartment, including dendritic cell commitment, inflammatory monocyte development, and eosinophil production. Many of these mouse phenotypes have been elegantly reproduced through analysis of human patients with inherited IRF8 mutations.7 One common thread throughout these pleiotropic effects is that IRF8 suppresses neutrophil potential,8 yet it is clear that IRF8 also promotes lineage-specific programs, such as GATA2, as shown by Sasaki and colleagues.1 How then does one transcription factor exert such diverse and cell-type–specific effects?
IRF8 and its closely related cousin, IRF4, possess relatively weak binding affinities for DNA.9 Thus, their major modes of action come from cooperating with other transcription factors to modify their activities and/or DNA binding specificities. Two major groups of partners for IRF4 and IRF8 are the Ets and AP-1 families of transcription factors.10 Using IRF8 point mutants, Sasaki and colleagues show that interaction with these partners is essential for promotion of the basophil lineage.1 However, many members of the Ets and AP-1 families are expressed during hematopoietic differentiation. Thus, it remains an open question which of these factors partners with IRF8 to drive basophil differentiation, and whether distinct partners are used for other lineages. Moreover, it remains unknown how IRF8 synergizes with other transcription factors, such as C/EBPα, which are also important for basophil commitment.4
Although many such questions still remain, Sasaki and colleagues have made a major advance by demonstrating the functional requirement for IRF8 in basophil development. By marrying modern cellular and genetic approaches, the authors bring some clarity to the steps underlying the development of this intriguing lineage.
Conflict-of-interest disclosure: The author declares no competing financial interests.