Key Points
In patients with previously diagnosed IPS, more than half (57%) had pathogens detected by currently available diagnostic methods.
Detection of a pathogen was significantly associated with high mortality regardless of significance of pathogenicity in lung.
Abstract
Newer diagnostic methods may link more idiopathic pneumonia syndrome (IPS) cases to an infectious agent. Bronchoalveolar lavage (BAL) samples from 69 hematopoietic cell transplant (HCT) recipients with IPS diagnosed between 1992 and 2006 were tested for 28 pathogens (3 bacteria and 25 viruses) by quantitative polymerase chain reaction and for Aspergillus by galactomannan assay. Research BALs from 21 asymptomatic HCT patients served as controls. Among 69 HCT patients with IPS, 39 (56.5%) had a pathogen detected. The most frequent pathogens were human herpesvirus-6 (HHV-6) (N = 20 [29%]) followed by human rhinovirus (HRV), cytomegalovirus (CMV), and Aspergillus (N = 8 [12%] in each). HHV-6 and HRV were rarely detected in controls, whereas CMV and Aspergillus were occasionally detected with low pathogen load. Patients with pathogens had worse day-100 survival than those without (hazard ratio, 1.88; P = .03). Mortality in patients with only pathogens of “uncertain” significance in lung was similar to that in patients with pathogens of “established” significance. Metagenomic next-generation sequencing did not reveal additional significant pathogens. Our study demonstrated that approximately half of patients with IPS had pathogens detected in BAL, and pathogen detection was associated with increased mortality. Thus, an expanded infection detection panel can significantly increase the diagnostic precision for idiopathic pneumonia.
Introduction
Idiopathic pneumonia syndrome (IPS) is a noninfectious pulmonary complication with diffuse lung injury after hematopoietic cell transplantation (HCT).1,2 According to previous reports, the incidence rate of IPS after allogeneic HCT is 4% to 12%, and the mortality from IPS remains high at 50% to 90%.3-12
Current diagnostic criteria of IPS are based on fulfilling 2 major requirements: (1) widespread alveolar injury with symptoms and signs of pneumonia and (2) absence of active lower respiratory tract infection.1,2 Detection of pathogens is a prerequisite to exclude infections. In the original diagnostic criteria, conventional or shell viral culture, direct fluorescent antibody testing, and pathological examination were used to detect pathogens.1 However, new diagnostic methods such as the biomarker galactomannan for Aspergillus and multiplex polymerase chain reaction (PCR) for respiratory viruses are now increasingly used. The recently modified diagnostic criteria suggest using these methods to diagnose IPS.2 Moreover, new pathogens, especially viruses such as human metapneumovirus (HMPV) or novel coronaviruses (CoVs), have been discovered using emerging technologies such as metagenomic next-generation sequencing (NGS).13,14
To examine our hypothesis that previously diagnosed IPS cases may include infectious pneumonia, we tested archived bronchoalveolar lavage (BAL) samples to identify occult pathogens using currently available techniques. Moreover, we evaluated the impact of these pathogens on mortality.
Methods
Study design
This study cohort includes patients who underwent HCT between 1992 and 2006 at the Fred Hutchinson Cancer Research Center (FHCRC) and had IPS within 120 days after transplantation,3 with an available BAL sample. To evaluate occult pathogens in asymptomatic control patients, we also studied patients transplanted between 1988 and 1991 at FHCRC who had no respiratory symptoms and a research BAL sample obtained between days 35 and 45 after HCT.15 The study was approved by the Institutional Review Board at FHCRC.
Laboratory testing
To detect pathogens using a BAL sample, the following tests were routinely performed until 2006 at our center: gram stain, fungal stain, and acid fast bacilli stain; cytology examination; conventional cultures for bacteria, mycobacteria, fungi, and viruses; shell viral culture for cytomegalovirus (CMV) and respiratory syncytial virus (RSV); and direct fluorescent antibody testing for Legionella, Pneumocystis jiroveci, CMV, RSV, parainfluenza virus (PIV) types 1 to 3, and adenovirus.16 For this study, BAL samples were further tested by real-time PCR and reverse transcription–PCR assays for the detection of 3 bacterial and 25 viral organisms: atypical bacteria (Legionella sp., Mycoplasma pneumoniae, and Chlamydia pneumoniae), herpesviruses (herpes simplex virus [HSV] types 1 and 2, varicella-zoster virus, Epstein-Barr virus [EBV], CMV, and human herpesvirus-6 [HHV-6]), polyomaviruses (PyVs; BK virus [BKV], JC virus, WUPyV, and KIPyV), adenovirus, parvovirus B19, enterovirus, parechoviruses, and respiratory viruses (RSV, PIV types 1-4, influenza types A and B, HMPV, human rhinoviruses [HRVs], human CoVs [OC43, 229E, NL63, and HKU1], and human bocavirus). Information about PCR analyses is provided in supplemental Table 1 (see the Blood Web site). Only samples with 2 positive results in duplicate assays were considered positive. For positive viral targets except HRV, pathogen load was determined by quantitative PCR, provided that sufficient specimen was available. High viral load was defined as >105 copies per mL or a PCR cycle threshold of ≤32 (when quantitation was not possible).
BAL and/or concurrent serum samples obtained between day −8 and day +6 at the time of BAL were examined for Aspergillus using the galactomannan assay. An index of ≥0.5 was considered positive.17
Sera obtained between day −8 and day +3 of the diagnosis were tested for the pathogen(s) detected in the BAL sample to determine the possibility of blood contamination of the BAL sample.
Follow-up BAL and/or autopsy samples obtained within 28 days after diagnosis of IPS were tested for any pathogen detected in the initial BAL sample to determine pathogen persistence and tissue invasion, respectively.
In patients whose BAL sample at diagnosis was positive for HHV-6, chromosomal integration of HHV-6 (ciHHV-6) was analyzed by droplet digital PCR using donor and/or recipient blood cells before HCT, if available.18
For NGS, 50 μL from 8 to 9 BAL samples was combined into individual pools and treated with DNase prior to nucleic acid extraction.19 NGS libraries were constructed using a modified Illumina TruSeq protocol as previously described.20 Samples were sequenced across 3 lanes of an Illumina HiSeq instrument. Analysis of NGS reads was performed using SURPI, a computational pipeline for pathogen detection that classifies NGS reads bioinformatically according to their origin (eg, human, bacteria, or virus).21
Definitions
Pathogens were categorized as “uncertain” if no consensus exists regarding their pathogenicity in lung.22-26 Diffuse alveolar hemorrhage (DAH) was confirmed by bronchoscopic findings.2 Death caused by respiratory failure was defined as any death caused exclusively or predominantly by respiratory failure.27
Statistical analysis
The probability of overall survival was estimated using the Kaplan-Meier method. The probability of mortality from respiratory failure was estimated by cumulative incidence curves, treating death because of other causes as a competing risk. The log-rank test was used to compare hazards of time-to-event outcomes among patients’ categories. Cox proportional hazards models were used to evaluate unadjusted and adjusted hazard ratios (aHRs) for mortality or respiratory mortality. Variables with P ≤ .1 in the univariable models were candidates for multivariable models. Two-sided P values <.05 were considered statistically significant. All statistical analyses were performed using SAS 9.3 for Windows (SAS Institute Inc., Cary, NC).
Results
Patient characteristics
A total of 69 patients diagnosed with IPS as well as 21 asymptomatic HCT control patients were evaluated. Characteristics of each group are shown in Table 1. The median age of patients with IPS was 46.1 years (range, 7-63). All patients in the control group underwent transplantation in the earlier years of the study period, received hematopoietic stem cells from bone marrow only, and were engrafted at the time of bronchoscopy. IPS cases without pathogen showed a trend toward being CMV seronegative (P = .055) and were more likely to have no or low-grade acute GVHD (P = .015).
Characteristics of patients with IPS and controls
| Characteristics . | Controls (N = 21) . | Patients with IPS . | ||
|---|---|---|---|---|
| Total (N = 69) . | Without pathogen (N = 31)* . | With pathogen (N = 38) . | ||
| Sex | ||||
| Male | 12 (57) | 35 (51) | 15 (48) | 20 (52) |
| Female | 9 (43) | 34 (49) | 16 (52) | 18 (48) |
| Age at transplantation, y | ||||
| ≤20 | 0 (0) | 4 (6) | 1 (3) | 3 (8) |
| 21-60 | 21 (100) | 63 (91) | 29 (94) | 34 (89) |
| >60 | 0 (0) | 2 (3) | 1 (3) | 1 (3) |
| Transplant year | ||||
| 1988-1998 | 21 (100) | 32 (46) | 12 (39) | 20 (53) |
| 1999-2006 | 0 (0) | 37 (54) | 19 (61) | 18 (47) |
| Disease risk at transplantation† | ||||
| Standard | 7 (33) | 37 (54) | 18 (58) | 19 (50) |
| High | 14 (67) | 32 (46) | 13 (42) | 19 (50) |
| Cell source | ||||
| Bone marrow | 21 (100) | 51 (74) | 22 (71) | 29 (76) |
| Peripheral blood stem cell | 0 (0) | 18 (26) | 9 (29) | 9 (24) |
| Donor type | ||||
| Matched related | 12 (57) | 23 (33) | 10 (32) | 13 (34) |
| Mismatched related | 2 (10) | 5 (7) | 3 (10) | 2 (5) |
| Unrelated | 7 (33) | 41 (59) | 18 (58) | 23 (61) |
| Conditioning regimen‡ | ||||
| MA including high-dose TBI | 18 (86) | 47 (68) | 20 (65) | 27 (71) |
| MA without TBI | 3 (14) | 21 (30) | 10 (32) | 11 (29) |
| Reduced intensity | 0 (0) | 1 (2) | 1 (3) | 0 (0) |
| GVHD prophylaxis | ||||
| CNI + MTX | 14 (67) | 62 (90) | 27 (87) | 35 (92) |
| Others§ | 7 (33) | 7 (10) | 4 (13) | 3 (8) |
| CMV serostatus | ||||
| Negative | 5 (24) | 33 (48) | 19 (61) | 14 (37) |
| Positive | 16 (76) | 36 (52) | 12 (39) | 24 (63) |
| % FEV1/FVC before transplantation | ||||
| ≥70 | 18 (86) | 55 (80) | 23 (74) | 32 (84) |
| <70 | 1 (5) | 11 (16) | 8 (26) | 3 (8) |
| Missing | 2 (10) | 3 (4) | 0 (0) | 3 (8) |
| % TLC before transplantation | ||||
| ≥80 | 19 (90) | 57 (83) | 28 (90) | 29 (76) |
| <80 | 0 (0) | 7 (10) | 3 (10) | 4 (11) |
| Missing | 2 (10) | 5 (7) | 0 (0) | 5 (13) |
| Days between transplantation and bronchoscopy | ||||
| ≤30 | 0 (0) | 48 (70) | 9 (29) | 12 (32) |
| 31-120 | 21 (100) | 21 (30) | 22 (71) | 26 (68) |
| Days between transplantation and bronchoscopy, median (range)|| | 42 (40-46) | 22 (4-119) | 19 (4-99) | 22.5 (8-119) |
| Bacteremia/candidemia at bronchoscopy | ||||
| No | 20 (95) | 64 (93) | 28 (90) | 36 (95) |
| Yes | 1 (5) | 4 (6) | 2 (6) | 2 (5) |
| Missing | 0 (0) | 1 (1) | 1 (3) | 0 (0) |
| White blood cell counts at bronchoscopy | ||||
| >1.0 × 10 e9/L | 21 (100) | 36 (52) | 15 (48) | 21 (55) |
| ≤1.0 × 10 e9/L | 0 (0) | 33 (48) | 16 (52) | 17 (45) |
| Lymphocyte count at bronchoscopy | ||||
| >0.2 × 10 e9/L | 17 (81) | 31 (45) | 13 (42) | 18 (47) |
| ≤0.2 × 10 e9/L | 4 (19) | 37 (54) | 17 (55) | 20 (53) |
| Missing | 0 (0) | 1 (1) | 1 (3) | 0 (0) |
| Neutrophil count at bronchoscopy | ||||
| >0.5 × 10 e9/L | 21 (100) | 37 (54) | 14 (45) | 23 (61) |
| ≤0.5 × 10 e9/L | 0 (0) | 32 (46) | 17 (55) | 15 (39) |
| Acute GVHD at bronchoscopy | ||||
| Grade 0-1 | 7 (33) | 20 (29) | 14 (45) | 6 (16) |
| Grade 2-4 | 14 (67) | 49 (71) | 17 (55) | 32 (84) |
| Steroid dose before bronchoscopy¶ | ||||
| No | 9 (43) | 37 (54) | 20 (64) | 17 (45) |
| <2 mg/kg | 5 (24) | 12 (17) | 4 (13) | 8 (21) |
| ≥2 mg/kg | 6 (29) | 20 (29) | 7 (23) | 13 (34) |
| Missing | 1 (5) | 0 (0) | 0 (0) | 0 (0) |
| DAH | ||||
| No | 21 (100) | 46 (67) | 23 (74) | 23 (61) |
| Yes | 0 (0) | 23 (33) | 8 (26) | 15 (39) |
| Treatment after bronchoscopy# | ||||
| None/steroid <1 mg/kg | 11 (52) | 8 (12) | 5 (16) | 3 (8) |
| Steroid 1-2 mg/kg | 9 (43) | 22 (32) | 11 (34) | 11 (29) |
| Steroid >2 mg/kg | 1 (5) | 39 (56) | 15 (48) | 24 (63) |
| TNF-receptor inhibitor | 0 (0) | 3 (4) | 3 (10) | 0 (0) |
| Characteristics . | Controls (N = 21) . | Patients with IPS . | ||
|---|---|---|---|---|
| Total (N = 69) . | Without pathogen (N = 31)* . | With pathogen (N = 38) . | ||
| Sex | ||||
| Male | 12 (57) | 35 (51) | 15 (48) | 20 (52) |
| Female | 9 (43) | 34 (49) | 16 (52) | 18 (48) |
| Age at transplantation, y | ||||
| ≤20 | 0 (0) | 4 (6) | 1 (3) | 3 (8) |
| 21-60 | 21 (100) | 63 (91) | 29 (94) | 34 (89) |
| >60 | 0 (0) | 2 (3) | 1 (3) | 1 (3) |
| Transplant year | ||||
| 1988-1998 | 21 (100) | 32 (46) | 12 (39) | 20 (53) |
| 1999-2006 | 0 (0) | 37 (54) | 19 (61) | 18 (47) |
| Disease risk at transplantation† | ||||
| Standard | 7 (33) | 37 (54) | 18 (58) | 19 (50) |
| High | 14 (67) | 32 (46) | 13 (42) | 19 (50) |
| Cell source | ||||
| Bone marrow | 21 (100) | 51 (74) | 22 (71) | 29 (76) |
| Peripheral blood stem cell | 0 (0) | 18 (26) | 9 (29) | 9 (24) |
| Donor type | ||||
| Matched related | 12 (57) | 23 (33) | 10 (32) | 13 (34) |
| Mismatched related | 2 (10) | 5 (7) | 3 (10) | 2 (5) |
| Unrelated | 7 (33) | 41 (59) | 18 (58) | 23 (61) |
| Conditioning regimen‡ | ||||
| MA including high-dose TBI | 18 (86) | 47 (68) | 20 (65) | 27 (71) |
| MA without TBI | 3 (14) | 21 (30) | 10 (32) | 11 (29) |
| Reduced intensity | 0 (0) | 1 (2) | 1 (3) | 0 (0) |
| GVHD prophylaxis | ||||
| CNI + MTX | 14 (67) | 62 (90) | 27 (87) | 35 (92) |
| Others§ | 7 (33) | 7 (10) | 4 (13) | 3 (8) |
| CMV serostatus | ||||
| Negative | 5 (24) | 33 (48) | 19 (61) | 14 (37) |
| Positive | 16 (76) | 36 (52) | 12 (39) | 24 (63) |
| % FEV1/FVC before transplantation | ||||
| ≥70 | 18 (86) | 55 (80) | 23 (74) | 32 (84) |
| <70 | 1 (5) | 11 (16) | 8 (26) | 3 (8) |
| Missing | 2 (10) | 3 (4) | 0 (0) | 3 (8) |
| % TLC before transplantation | ||||
| ≥80 | 19 (90) | 57 (83) | 28 (90) | 29 (76) |
| <80 | 0 (0) | 7 (10) | 3 (10) | 4 (11) |
| Missing | 2 (10) | 5 (7) | 0 (0) | 5 (13) |
| Days between transplantation and bronchoscopy | ||||
| ≤30 | 0 (0) | 48 (70) | 9 (29) | 12 (32) |
| 31-120 | 21 (100) | 21 (30) | 22 (71) | 26 (68) |
| Days between transplantation and bronchoscopy, median (range)|| | 42 (40-46) | 22 (4-119) | 19 (4-99) | 22.5 (8-119) |
| Bacteremia/candidemia at bronchoscopy | ||||
| No | 20 (95) | 64 (93) | 28 (90) | 36 (95) |
| Yes | 1 (5) | 4 (6) | 2 (6) | 2 (5) |
| Missing | 0 (0) | 1 (1) | 1 (3) | 0 (0) |
| White blood cell counts at bronchoscopy | ||||
| >1.0 × 10 e9/L | 21 (100) | 36 (52) | 15 (48) | 21 (55) |
| ≤1.0 × 10 e9/L | 0 (0) | 33 (48) | 16 (52) | 17 (45) |
| Lymphocyte count at bronchoscopy | ||||
| >0.2 × 10 e9/L | 17 (81) | 31 (45) | 13 (42) | 18 (47) |
| ≤0.2 × 10 e9/L | 4 (19) | 37 (54) | 17 (55) | 20 (53) |
| Missing | 0 (0) | 1 (1) | 1 (3) | 0 (0) |
| Neutrophil count at bronchoscopy | ||||
| >0.5 × 10 e9/L | 21 (100) | 37 (54) | 14 (45) | 23 (61) |
| ≤0.5 × 10 e9/L | 0 (0) | 32 (46) | 17 (55) | 15 (39) |
| Acute GVHD at bronchoscopy | ||||
| Grade 0-1 | 7 (33) | 20 (29) | 14 (45) | 6 (16) |
| Grade 2-4 | 14 (67) | 49 (71) | 17 (55) | 32 (84) |
| Steroid dose before bronchoscopy¶ | ||||
| No | 9 (43) | 37 (54) | 20 (64) | 17 (45) |
| <2 mg/kg | 5 (24) | 12 (17) | 4 (13) | 8 (21) |
| ≥2 mg/kg | 6 (29) | 20 (29) | 7 (23) | 13 (34) |
| Missing | 1 (5) | 0 (0) | 0 (0) | 0 (0) |
| DAH | ||||
| No | 21 (100) | 46 (67) | 23 (74) | 23 (61) |
| Yes | 0 (0) | 23 (33) | 8 (26) | 15 (39) |
| Treatment after bronchoscopy# | ||||
| None/steroid <1 mg/kg | 11 (52) | 8 (12) | 5 (16) | 3 (8) |
| Steroid 1-2 mg/kg | 9 (43) | 22 (32) | 11 (34) | 11 (29) |
| Steroid >2 mg/kg | 1 (5) | 39 (56) | 15 (48) | 24 (63) |
| TNF-receptor inhibitor | 0 (0) | 3 (4) | 3 (10) | 0 (0) |
All values are indicated as the number (percentage).
CNI, calcineurin inhibitor; CSP, cyclosporine; FEV1, forced expiratory volume in 1 s; FVC, forced vital capacity; GVHD, graft-versus-host disease; MA, myeloablative; MMF, mycophenolate mofetil; MTX, methotorexate; TBI, total body irradiation; % TLC, percentage of predicted total lung capacity; TNF, tumor necrosis factor.
This includes a patient with ciHHV-6.
Disease risk group at transplantation was classified into 2 groups: standard and high as previously described.27
Myeloablative conditioning regimens mainly consisted of high-dose cyclophosphamide and busulfan with or without fractionated TBI (12.0 or 13.2 Gy). Reduced intensity conditioning regimen consisted of fludarabine with a single fraction of TBI (2 Gy).
Others include MTX alone, CSP alone, CSP + steroid, and CSP + MMF.
This variable indicates median (range).
Steroid dose before diagnosis of IPS was defined as peak dose from the period within 2 weeks before bronchoscopy.38
The treatment was started within 3 weeks after bronchoscopy. Steroid dose was defined as peak dose within 3 weeks after bronchoscopy. Three patients that received TNF-receptor inhibitor were also included in the group of steroid >2 mg/kg.
Detection of occult pathogens
Among patients with IPS, 39 (56.5%) had occult pathogens (Table 2) and 16 had multiple ones (2 in 14 and >3 in 2). Of the 28 pathogens that were examined, 13 were detected at least once. We divided the 13 pathogens into 2 groups according to pathogenicity in lung: “established” and “uncertain” (Table 2). Aspergillus and CMV were the most common established pathogens, detected in 12% of the patients with IPS. HHV-6 was the most common pathogen among the uncertain group, detected in 20 patients (29%), 4 of whom had possible HHV-6–associated encephalitis (1 had HHV-6 DNA detected in cerebrospinal fluid). ciHHV-6 was observed in 1 of 17 patients with available cellular blood cell samples from donor and/or recipient before HCT (data not shown). The second most common uncertain pathogen was HRV, which frequently presented with high viral load. Among 18 patients with uncertain pathogens alone, all but 2 had HHV-6 and/or HRV, and HSV and EBV were each detected in 1 patient. In the control group, Aspergillus and CMV were also frequently detected, whereas HHV-6 and HRV were rarely detected. KIPyV was also detected in control patients with median viral load of 1.7 × 104 copies per mL (range, 3.0 × 103 to 3.1 × 104); 1 of the 5 KIPyV-positive patients had concomitant detection in plasma (6.2 × 102 copies per mL). Overall, only 1 of the samples from the controls showed high pathogen load.
Detected pathogens in BAL samples at diagnosis
| Pathogenicity in lung . | Detected pathogens . | IPS cases (N = 69) . | Controls (N = 21) . | P† . | ||
|---|---|---|---|---|---|---|
| Pathogen load . | Pathogen load . | |||||
| Any (N = 39) . | High* (N = 12) . | Any (N = 13) . | High* (N = 1) . | |||
| Established | Aspergillus‡ | 8 (12) | N/A | 3 (14) | N/A | .71 |
| CMV | 8 (12) | 0 (0) | 6 (29) | 0 (0) | .08 | |
| PIV | 2 (3) | 0 (0) | 0 (0) | 0 (0) | 1.00 | |
| RSV | 2 (3) | 1 (1) | 0 (0) | 0 (0) | 1.00 | |
| HMPV | 2 (3) | 2 (3) | 0 (0) | 0 (0) | 1.00 | |
| Uncertain | HHV-6 | 20 (29) | 4 (6) | 1 (5) | 0 (0) | .020 |
| HRV | 8 (12) | 6 (9) | 1 (5) | 0 (0) | .68 | |
| KIPyV | 3 (4) | 1 (1) | 5 (24) | 0 (0) | .016 | |
| BKV | 3 (4) | 0 (0) | 0 (0) | 0 (0) | 1.00 | |
| HSV | 2 (3) | 0 (0) | 0 (0) | 0 (0) | 1.00 | |
| EBV | 1 (1) | 0 (0) | 2 (10) | 0 (0) | .14 | |
| WUPyV | 1 (1) | 0 (0) | 0 (0) | 0 (0) | 1.00 | |
| CoV | 0 (0) | 0 (0) | 1 (5) | 1 (5) | .23 | |
| Pathogenicity in lung . | Detected pathogens . | IPS cases (N = 69) . | Controls (N = 21) . | P† . | ||
|---|---|---|---|---|---|---|
| Pathogen load . | Pathogen load . | |||||
| Any (N = 39) . | High* (N = 12) . | Any (N = 13) . | High* (N = 1) . | |||
| Established | Aspergillus‡ | 8 (12) | N/A | 3 (14) | N/A | .71 |
| CMV | 8 (12) | 0 (0) | 6 (29) | 0 (0) | .08 | |
| PIV | 2 (3) | 0 (0) | 0 (0) | 0 (0) | 1.00 | |
| RSV | 2 (3) | 1 (1) | 0 (0) | 0 (0) | 1.00 | |
| HMPV | 2 (3) | 2 (3) | 0 (0) | 0 (0) | 1.00 | |
| Uncertain | HHV-6 | 20 (29) | 4 (6) | 1 (5) | 0 (0) | .020 |
| HRV | 8 (12) | 6 (9) | 1 (5) | 0 (0) | .68 | |
| KIPyV | 3 (4) | 1 (1) | 5 (24) | 0 (0) | .016 | |
| BKV | 3 (4) | 0 (0) | 0 (0) | 0 (0) | 1.00 | |
| HSV | 2 (3) | 0 (0) | 0 (0) | 0 (0) | 1.00 | |
| EBV | 1 (1) | 0 (0) | 2 (10) | 0 (0) | .14 | |
| WUPyV | 1 (1) | 0 (0) | 0 (0) | 0 (0) | 1.00 | |
| CoV | 0 (0) | 0 (0) | 1 (5) | 1 (5) | .23 | |
All values are indicated as the number of cases (percentage of cases with pathogens).
N/A, not applicable.
High load: >105 copies per mL, <32 cycle threshold (for HRV).
The values were obtained from the comparison in the number of patients with any pathogens between IPS cases and controls.
The median values of galactomannan were 0.65 (range, 0.57-2.37) in IPS cases and 1.46 (range, 0.62-2.24) in controls.
To detect occult pathogens, unbiased metagenomic NGS was also performed. Among 707 663 254 total reads, 27 080 corresponded to significant viral pathogens (supplemental Table 2 and Table 3). Notably, no additional pathogens beyond those detected by PCR testing were observed except hepatitis C virus in 1 pool that included a virus carrier (Table 3).
Verified viral summary
| Samples . | Verified viral signature . | Reads identified . | % Target reads . | % Genomic coverage . |
|---|---|---|---|---|
| BAL_Pool_1 | HHV-6 | 805 | 0.0013 | 9.6 |
| Rhinovirus B | 150 | 0.00023 | 17.0 | |
| Hepatitis C virus | 38 | 0.000059 | 10.7 | |
| Bovine viral diarrhea virus 1* | 64 | 0.00010 | 7.5 | |
| Anelloviridae† | 23 | 0.00000036 | N/A | |
| BAL_Pool_2 | RSV | 228 | 0.00033 | 45.0 |
| Rhinovirus B | 115 | 0.00017 | 59.5 | |
| HHV-6 | 14 | 0.000020 | 0.8 | |
| CMV | 2 | 0.0000029 | 0.1 | |
| Bovine viral diarrhea virus 1* | 3 | 0.0000044 | 2.2 | |
| BAL_Pool_3 | HHV-6 | 348 | 0.00054 | 10.5 |
| Rhinovirus B | 1 | 0.0000015 | 1.4 | |
| Bovine viral diarrhea virus 1* | 37 | 0.000057 | 12.3 | |
| Anelloviridae† | 8353 | 0.00013 | N/A | |
| BAL_Pool_4 | HHV- 6 | 33 | 0.000048 | 1.2 |
| Bovine viral diarrhea virus 1* | 21 | 0.000031 | 12.8 | |
| Anelloviridae† | 306 | 0.0000045 | N/A | |
| BAL_Pool_5 | Rhinovirus B | 8 | 0.000011 | 5.2 |
| CMV | 1 | 0.0000013 | 0.0 | |
| Anelloviridae† | 3 | 0.00000004 | N/A | |
| BAL_Pool_6 | HHV-6 | 698 | 0.0011 | 24.9 |
| Rhinovirus A | 260 | 0.00040 | 76.7 | |
| Rhinovirus B | 82 | 0.00013 | 39.8 | |
| KIPyV | 5 | 0.0000077 | 9.2 | |
| CMV | 2 | 0.0000031 | 0.1 | |
| Bovine viral diarrhea virus 1* | 2 | 0.0000031 | 1.6 | |
| Anelloviridae† | 234 | 0.0000036 | N/A | |
| BAL_Pool_7 | HMPV | 104 | 0.00010 | 27.5 |
| HHV-6 | 37 | 0.030036 | 1.6 | |
| Bovine viral diarrhea virus 1* | 13 | 0.000013 | 6.6 | |
| Anelloviridae† | 17 632 | 0.00017 | N/A | |
| BAL_Pool_8 | HMPV | 24 062 | 0.021 | 99.4 |
| HHV-6 | 51 | 0.000045 | 1.7 | |
| Anelloviridae† | 219 | 0.0000019 | N/A |
| Samples . | Verified viral signature . | Reads identified . | % Target reads . | % Genomic coverage . |
|---|---|---|---|---|
| BAL_Pool_1 | HHV-6 | 805 | 0.0013 | 9.6 |
| Rhinovirus B | 150 | 0.00023 | 17.0 | |
| Hepatitis C virus | 38 | 0.000059 | 10.7 | |
| Bovine viral diarrhea virus 1* | 64 | 0.00010 | 7.5 | |
| Anelloviridae† | 23 | 0.00000036 | N/A | |
| BAL_Pool_2 | RSV | 228 | 0.00033 | 45.0 |
| Rhinovirus B | 115 | 0.00017 | 59.5 | |
| HHV-6 | 14 | 0.000020 | 0.8 | |
| CMV | 2 | 0.0000029 | 0.1 | |
| Bovine viral diarrhea virus 1* | 3 | 0.0000044 | 2.2 | |
| BAL_Pool_3 | HHV-6 | 348 | 0.00054 | 10.5 |
| Rhinovirus B | 1 | 0.0000015 | 1.4 | |
| Bovine viral diarrhea virus 1* | 37 | 0.000057 | 12.3 | |
| Anelloviridae† | 8353 | 0.00013 | N/A | |
| BAL_Pool_4 | HHV- 6 | 33 | 0.000048 | 1.2 |
| Bovine viral diarrhea virus 1* | 21 | 0.000031 | 12.8 | |
| Anelloviridae† | 306 | 0.0000045 | N/A | |
| BAL_Pool_5 | Rhinovirus B | 8 | 0.000011 | 5.2 |
| CMV | 1 | 0.0000013 | 0.0 | |
| Anelloviridae† | 3 | 0.00000004 | N/A | |
| BAL_Pool_6 | HHV-6 | 698 | 0.0011 | 24.9 |
| Rhinovirus A | 260 | 0.00040 | 76.7 | |
| Rhinovirus B | 82 | 0.00013 | 39.8 | |
| KIPyV | 5 | 0.0000077 | 9.2 | |
| CMV | 2 | 0.0000031 | 0.1 | |
| Bovine viral diarrhea virus 1* | 2 | 0.0000031 | 1.6 | |
| Anelloviridae† | 234 | 0.0000036 | N/A | |
| BAL_Pool_7 | HMPV | 104 | 0.00010 | 27.5 |
| HHV-6 | 37 | 0.030036 | 1.6 | |
| Bovine viral diarrhea virus 1* | 13 | 0.000013 | 6.6 | |
| Anelloviridae† | 17 632 | 0.00017 | N/A | |
| BAL_Pool_8 | HMPV | 24 062 | 0.021 | 99.4 |
| HHV-6 | 51 | 0.000045 | 1.7 | |
| Anelloviridae† | 219 | 0.0000019 | N/A |
This is contamination of the sample of the libraries.
This is nonpathological flora.
Confirmation of detected pathogens
Among 38 IPS cases with pathogen, excluding the 1 ciHHV-6 case, 16 had a follow-up BAL and/or autopsy sample within 28 days after diagnosis (supplemental Table 4). The same pathogens detected at diagnosis were again detected in all but 2 cases, and for 8 of 11 pathogens, pathogen loads were stable or increased.
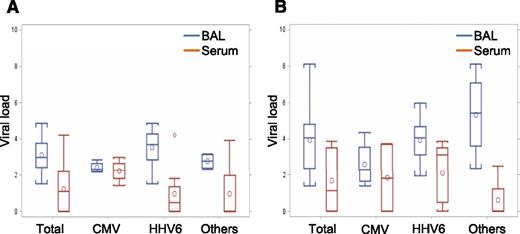
To evaluate whether the viruses in BAL are a reflection of contaminated blood, we compared viral loads between BAL and concurrently obtained serum in both DAH-positive cases (20 tests in 12 cases) (Figure 1A) and DAH-negative cases (19 tests in 18 cases) (Figure 1B). Only HHV-6 (N = 11), CMV (N = 6), BKV (N = 1), and HMPV (N = 1) were detected in sera (supplemental Table 5), and no viruses except CMV in 4 out of 6 cases were detected in sera during the clinical time course. Most cases had higher viral load in BAL than serum, and only 3 cases with CMV and 1 case with BKV showed higher viral load in serum than in BAL.
Comparison of viral load between BAL and serum. (A) Viral load in samples from patients with DAH (N = 20). Each group indicates viral load in BAL (blue box) and serum (red box). P values are .002, .88, .002, and .30 in total, CMV, HHV-6, and others, respectively. (B) Viral load in samples from patients without DAH (N = 19). Each group indicates viral titer in BAL (blue box) and serum (red box). P values are .003, .66, .030, and .055 in total, CMV, HHV-6, and others, respectively.
Comparison of viral load between BAL and serum. (A) Viral load in samples from patients with DAH (N = 20). Each group indicates viral load in BAL (blue box) and serum (red box). P values are .002, .88, .002, and .30 in total, CMV, HHV-6, and others, respectively. (B) Viral load in samples from patients without DAH (N = 19). Each group indicates viral titer in BAL (blue box) and serum (red box). P values are .003, .66, .030, and .055 in total, CMV, HHV-6, and others, respectively.
Mortality after IPS by presence of pathogen
Among the 69 patients with IPS, 38 patients had occult pathogens: 20 patients with established pathogens and 18 with uncertain pathogen alone. One patient with ciHHV-6 was categorized as without pathogen.
Overall survival by 100 days in IPS patients with pathogens was significantly worse than that in patients without pathogens (P < .0001; Figure 2A). Similar results were obtained in mortality from respiratory failure (P < .0001; Figure 2B). This difference persisted for 5 years. Transplant year did not affect the mortality. In a multivariable analysis, detection of pathogen was significantly associated with high mortality among patients with IPS (aHR, 2.08; 95% confidence interval [CI], 1.16-3.34; P = .03) (Table 4). For respiratory death, detection of a pathogen was also an independently important factor (aHR, 2.28; 95% CI, 1.15-4.52; P = .02) (Table 4).
Probability of overall survival and respiratory death by presence of pathogens. (A) Kaplan-Meier estimate of overall survival by presence of pathogens. One patient with ciHHV-6 was included in the IPS cases without pathogens. (B) Cumulative incidence of respiratory death by presence of pathogens. (C) Kaplan-Meier estimate of overall survival by pathogen type (aHR for established vs uncertain, 0.83 [0.41-1.69]; P = .61). (D) Cumulative incidence of respiratory death by pathogen type (aHR for established vs uncertain, 0.73 [0.33-1.61]; P = .44).
Probability of overall survival and respiratory death by presence of pathogens. (A) Kaplan-Meier estimate of overall survival by presence of pathogens. One patient with ciHHV-6 was included in the IPS cases without pathogens. (B) Cumulative incidence of respiratory death by presence of pathogens. (C) Kaplan-Meier estimate of overall survival by pathogen type (aHR for established vs uncertain, 0.83 [0.41-1.69]; P = .61). (D) Cumulative incidence of respiratory death by pathogen type (aHR for established vs uncertain, 0.73 [0.33-1.61]; P = .44).
Risk factors for mortality from all causes or respiratory failure by day 100 after diagnosis of IPS, N = 69
| . | Univariable analysis . | Multivariable analysis . | ||||
|---|---|---|---|---|---|---|
| HR . | 95% CI . | P . | HR . | 95% CI . | P . | |
| Overall mortality | ||||||
| Donor type | ||||||
| Matched related | 1.00 | 1.00 | ||||
| Mismatched related/unrelated | 0.48 | 0.28-0.84 | .01 | 0.54 | 0.30-0.95 | .03 |
| Bacteremia/candidemia | ||||||
| No | 1.00 | 1.00 | ||||
| Yes | 2.59 | 0.92-7.31 | .07 | 3.09 | 1.06-9.04 | .04 |
| Steroid dose before diagnosis | ||||||
| No | 1.00 | 1.00 | ||||
| ≤1 mg/kg | 2.25 | 1.09-4.65 | .03 | 1.75 | 0.99-3.10 | .05 |
| >1 mg/kg | 2.13 | 1.15-3.95 | .02 | |||
| Steroid dose after diagnosis* | ||||||
| ≤2 mg/kg | 1.00 | 1.00 | ||||
| >2 mg/kg | 2.86 | 1.07-7.63 | .04 | 3.56 | 1.18-10.77 | .02 |
| DAH | ||||||
| No | 1.00 | |||||
| Yes | 1.73 | 0.98-3.06 | .06 | |||
| Detected pathogen | ||||||
| None | 1.00 | 1.00 | ||||
| Established | 1.80 | 0.94-3.43 | .08 | 1.88 | 1.06-3.34 | .03 |
| Uncertain | 1.78 | 0.92-3.47 | .09 | |||
| Viral load† | ||||||
| Low | 1.00 | |||||
| High | 0.71 | 0.32-1.55 | .38 | |||
| Mortality from respiratory failure | ||||||
| Donor type | ||||||
| Matched related | 1.00 | 1.00 | ||||
| Mismatched related/unrelated | 0.48 | 0.26-0.91 | .02 | 0.54 | 0.29-1.04 | .06 |
| Bacteremia/candidemia | ||||||
| No | 1.00 | 1.00 | ||||
| Yes | 3.11 | 1.09-8.88 | .03 | 3.76 | 1.25-11.37 | .02 |
| Steroid dose before diagnosis | ||||||
| No | 1.00 | 1.00 | ||||
| ≤1 mg/kg | 2.99 | 1.33-6.71 | .008 | 2.20 | 1.12-4.29 | .02 |
| >1 mg/kg | 2.84 | 1.40-5.77 | .004 | |||
| Steroid dose after diagnosis* | ||||||
| ≤2 mg/kg | 1.00 | 1.00 | ||||
| >2 mg/kg | 4.49 | 1.29-15.63 | .02 | 6.83 | 1.50-31.04 | .01 |
| DAH | ||||||
| No | 1.00 | |||||
| Yes | 2.59 | 1.39-4.83 | .003 | |||
| Detected pathogen | ||||||
| None | 1.00 | 1.00 | ||||
| Established | 2.09 | 0.99-4.43 | .05 | 2.28 | 1.15-4.52 | .02 |
| Uncertain | 2.24 | 1.05-4.79 | .04 | |||
| Viral load† | ||||||
| Low | 1.00 | |||||
| High | 0.50 | 0.20-1.26 | .14 | |||
| . | Univariable analysis . | Multivariable analysis . | ||||
|---|---|---|---|---|---|---|
| HR . | 95% CI . | P . | HR . | 95% CI . | P . | |
| Overall mortality | ||||||
| Donor type | ||||||
| Matched related | 1.00 | 1.00 | ||||
| Mismatched related/unrelated | 0.48 | 0.28-0.84 | .01 | 0.54 | 0.30-0.95 | .03 |
| Bacteremia/candidemia | ||||||
| No | 1.00 | 1.00 | ||||
| Yes | 2.59 | 0.92-7.31 | .07 | 3.09 | 1.06-9.04 | .04 |
| Steroid dose before diagnosis | ||||||
| No | 1.00 | 1.00 | ||||
| ≤1 mg/kg | 2.25 | 1.09-4.65 | .03 | 1.75 | 0.99-3.10 | .05 |
| >1 mg/kg | 2.13 | 1.15-3.95 | .02 | |||
| Steroid dose after diagnosis* | ||||||
| ≤2 mg/kg | 1.00 | 1.00 | ||||
| >2 mg/kg | 2.86 | 1.07-7.63 | .04 | 3.56 | 1.18-10.77 | .02 |
| DAH | ||||||
| No | 1.00 | |||||
| Yes | 1.73 | 0.98-3.06 | .06 | |||
| Detected pathogen | ||||||
| None | 1.00 | 1.00 | ||||
| Established | 1.80 | 0.94-3.43 | .08 | 1.88 | 1.06-3.34 | .03 |
| Uncertain | 1.78 | 0.92-3.47 | .09 | |||
| Viral load† | ||||||
| Low | 1.00 | |||||
| High | 0.71 | 0.32-1.55 | .38 | |||
| Mortality from respiratory failure | ||||||
| Donor type | ||||||
| Matched related | 1.00 | 1.00 | ||||
| Mismatched related/unrelated | 0.48 | 0.26-0.91 | .02 | 0.54 | 0.29-1.04 | .06 |
| Bacteremia/candidemia | ||||||
| No | 1.00 | 1.00 | ||||
| Yes | 3.11 | 1.09-8.88 | .03 | 3.76 | 1.25-11.37 | .02 |
| Steroid dose before diagnosis | ||||||
| No | 1.00 | 1.00 | ||||
| ≤1 mg/kg | 2.99 | 1.33-6.71 | .008 | 2.20 | 1.12-4.29 | .02 |
| >1 mg/kg | 2.84 | 1.40-5.77 | .004 | |||
| Steroid dose after diagnosis* | ||||||
| ≤2 mg/kg | 1.00 | 1.00 | ||||
| >2 mg/kg | 4.49 | 1.29-15.63 | .02 | 6.83 | 1.50-31.04 | .01 |
| DAH | ||||||
| No | 1.00 | |||||
| Yes | 2.59 | 1.39-4.83 | .003 | |||
| Detected pathogen | ||||||
| None | 1.00 | 1.00 | ||||
| Established | 2.09 | 0.99-4.43 | .05 | 2.28 | 1.15-4.52 | .02 |
| Uncertain | 2.24 | 1.05-4.79 | .04 | |||
| Viral load† | ||||||
| Low | 1.00 | |||||
| High | 0.50 | 0.20-1.26 | .14 | |||
All variables in Table 1 were used for the univariable analysis. Only variables with P < .1 are shown in this table. Viral load was shown regardless of P values.
HR, hazard ratio.
Peak steroid doses after diagnosis and the peak date were recorded from within 2 weeks after diagnosis.38 These variables are analyzed as time dependent.
This analysis was performed only among 38 patients with viral pathogens.
Effect of pathogen type and viral load on mortality
A comparison between uncertain and established pathogens on mortality failed to show a significant effect of known pathogenicity (Figure 2C-D; Table 4). Among patients with uncertain pathogen, the survival in only patients with HHV-6 and/or HRV was also similar to that in patients with established pathogen (supplemental Figure 1). Among 36 patients with detected viruses, we compared mortality after IPS with the level of viral load. Low viral load had no survival advantage in both overall death and respiratory death (Table 4).
Discussion
This study demonstrates that more than half of the patients with previously diagnosed IPS had occult pathogens using currently available diagnostic methods and that detection of pathogens is significantly associated with high mortality regardless of known pathogenicity in lung.
Although many studies on incidence, risk factors, or outcome of IPS have been reported, the strategies of exclusion of infectious pneumonia vary (supplemental Table 6).3-12 All studies, including those conducted at our center, are based on the classical diagnostic criteria published by a National Institutes of Health workshop in 1993,1,3,8 and most of the studies used only cultures and stains to detect pathogens. Because the absence of lower respiratory tract infection is an essential component of the diagnosis of IPS, the diagnostic precision depends on the diagnostic panel used on the BAL fluid. Aspergillus galactomannan testing and multiplex respiratory virus testing have become more prevalent in recent years. However, these tests are not uniformly done by all centers for BAL workup. Recent diagnostic criteria by the American Thoracic Society in 2011 have not yet been fully implemented, and even in a recently published prospective study shown in supplemental Table 6, the microbial testing to diagnose IPS was insufficient. Using an extensive panel in this study, we found that more than half of the patients with previously diagnosed IPS had a pathogen detected and ∼30% of the patients had a pathogen that is known to cause pneumonia.
A key question in interpreting the results relates to the relevance of the detected organisms. To address this question, we divided pathogens into those with established pathogenicity in the lung including CMV, PIV, RSV, HMPV, and Aspergillus, and those of uncertain or poorly established causes of pneumonia (Table 2). Of the established pathogens, the respiratory viruses and Aspergillus are now readily detectable by multiplex PCR and galactomannan testing, respectively. Our data indicate their significance and support the recently modified diagnostic criteria reported by American Thoracic Society, which include these diagnostic methods to rule out infection during workup of potential IPS cases.2
CMV was the most commonly detected established pathogen. As expected, low-level CMV DNA in the BAL was detected in both cases and controls, which likely represents pulmonary CMV shedding rather than invasive disease. Asymptomatic shedding of CMV is a well-established phenomenon in HCT recipients that occurs in approximately one-third of seropositive recipients.15,28 It is per se not indicative of CMV pneumonia but constitutes a risk factor for subsequent progression to pneumonia when left untreated.28 This is consistent with our finding that persistent CMV DNA detection in 1 follow-up BAL and 2 autopsy samples from IPS cases did not result in CMV pneumonia (supplemental Table 4), whereas 4 control patients who had not received preemptive therapy died of CMV pneumonia (Figure 2D). There is no established viral load threshold that differentiates CMV pulmonary shedding from invasive disease.
Aspergillus galactomannan was also detected in >10% of IPS cases or controls. Most patients with IPS received anti-Aspergillus agents, whereas control patients did not. The 3 control patients with a positive galactomannan test did not progress to Aspergillus pneumonia despite a lack of treatment. These findings suggest that Aspergillus galactomannan can be detected in asymptomatic HCT patients after engraftment. Further studies are needed to define galactomannan thresholds and the role of presence or absence of neutropenia in asymptomatic HCT recipients.
Among the pathogens of uncertain pulmonary pathogenicity, HHV-6 and HRV were most frequently detected. Several lines of evidence support the importance of these pathogens. First, similarly high mortality rates were observed in patients with “established” and “uncertain” pathogens regardless of pathogenicity in lung (Figure 2C-D). Although both HHV-6 and HRV have been suggested to be associated with pneumonia after HCT in previous reports,25,29-33 we deliberately categorized them into “uncertain” pathogens because the published literature is inconclusive and no consensus exists.22-26 We found that ∼90% of the patients with uncertain pathogens had HHV-6 and/or HRV and that their outcome was as poor as that with established pathogens (supplemental Figure 1). Second, follow-up BAL and/or autopsy samples were available in ∼40% of the patients, and we detected the original pathogen in most of the follow-up samples, often with increased or persistent viral load, and even in autopsy samples (supplemental Table 4). These findings argue against temporary self-limited infection and support a pathogenic role of HHV-6 and HRV. Testing for HHV-6 and HRV should be considered as recommended by the recently modified diagnostic criteria of the American Thoracic Society,2 but studies are needed to validate this finding for the significance of HHV-6 or HRV in lung.
To further establish the significance of the detected organisms, we analyzed the impact of pathogen load on outcome. Pathogen burden overall was, somewhat surprisingly, not associated with outcome, possibly because of small sample size and nonstandardized quantitation in this retrospective study, which did not allow us to adjust for the dilution of the BAL fluid. There are presently no established thresholds for specific pathogens that are associated with pneumonia, and it is presently unknown whether low viral load can cause lung injury. This is a fundamental question concerning the significance of viral load in the development of pulmonary tissue pathology and clinical pneumonia. Studies are needed to define thresholds of viral load and specific etiologies of pneumonia. Among patients with pathogens, 42% had multiple pathogens detected, and most pathogens were continuously detected (supplemental Table 4). It may be difficult to attribute outcome to 1 particular pathogen, which is a common dilemma in infectious pneumonia in immunocompromised patients.
In addition to a comprehensive panel, we performed pathogen discovery and surveillance studies using metagenomic NGS to determine whether additional known, uncommon, or novel viral pathogens could be detected. This strategy has been previously shown to be effective in detection of bacteria19 and viruses20 in clinical samples with sensitivity comparable to specific PCR (supplemental Figure 2). Importantly, the NGS studies did not reveal any additional known or novel pathogens that were not already detected by specific viral PCR (Table 3). Bovine viral diarrhea virus is likely a laboratory reagent contaminant from fetal bovine serum. Viruses corresponding to the family, Anelloviridae, were also seen in 7 of 8 pools. These genetically diverse viruses have never been linked to disease and are considered to be part of the nonpathogenic flora.34 Nevertheless, it has been suggested that temporal fluctuations in the levels of Anelloviruses may serve as markers of immunosuppression in transplant recipients35 as well as lung inflammation.36 Although the limits of detection of NGS for broad-spectrum identification of infectious agents have yet to be established, these NGS findings suggest that a significant fraction of IPS cases may be because of noninfectious causes.
As for the association between pathogen detection and outcomes, we found that patients with pathogens had worse survival than patients without (Figure 2A-B). Considering that standard treatment of IPS is administration of high-dose corticosteroids,37 this result is reasonable because of the known negative impact of high-dose steroids on infectious pneumonia.38-41 This finding also indicates that strict diagnostic criteria may identify cases of “true” IPS, which will be critical for the interpretation of outcome studies and the design of future interventional strategies.
This study has strengths and limitations. This represents the largest cohort of IPS patients studied with a broad range of diagnostic tests and platforms using prospectively collected BAL, blood, and lung tissue samples. This study also included a control cohort of asymptomatic HCT recipients who underwent a research BAL and carefully accounted for the possible effect of pulmonary hemorrhage as a confounder. However, limitations should be noted. First, although we tried to identify occult pathogens using quantitative PCR and metagenomic NGS, we may have missed some pathogens, especially those present at low titers. The sensitivity of pathogen detection using NGS may be reduced by the pooled sample approach, and DNase digestion prior to extraction to reduce host background may have reduced sensitivity of their detection. However, we believe that the significance of potentially missed low-level novel infection, especially from uncommon or novel agents, would have been difficult to prove. Another limitation is a small sample size, although the number is substantial compared with other reports (supplemental Table 6). We also could not analyze the effect of pathogen type by each uncertain pathogen in a multivariable analysis, and the impact of pathogen load on mortality remains unclear. Finally, the control group had limitations, including an earlier era of transplantation, a narrower window and later time after HCT, and a lack of patients undergoing peripheral blood stem cell transplantation. These differences can affect detected pathogens or outcomes.
In conclusion, our findings show that approximately half of the patients with previously diagnosed IPS had occult pathogens and the presence of pathogens was associated with worse survival, even for pathogens whose pathogenicity in lung has not been firmly established. Thus, adding high-dose steroids in patients with detectable pathogens may be harmful and should be carefully considered. Furthermore, NGS did not seem to detect a significant number of pathogens that more readily accessible diagnostics cannot identify. Our data provide strong support for bronchoscopic examination and an intensive testing of pathogens in patients who present with lower respiratory tract disease to prevent misclassification of the disease and mistreatment in HCT recipients with IPS. Moreover, the results of this study may also be relevant to patients with acute lung injury after chemotherapy or immunotherapy for hematologic malignancies. We identified new viruses as potential pulmonary pathogens in this study, which will impact both daily clinical work and future studies. Further studies are required to confirm the effect of pathogen type and load in order to develop optimized treatment of IPS.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Zachary Stednick for database services; Terry Stevens-Ayers, Jessica N. Yi, Tracy K. Santo, and Linda Cook for laboratory assistance; Amanda C. Moklebust for preparation of pathology samples; Edith Rutledge and Bernadette Poydras for preparation of medical charts; and Raleigh Bowden for cryopreserving the BAL fluid samples from controls.
This work was partially supported by grants from the National Institutes of Health, National Heart, Lung and Blood Institute (HL081595 and K24HL093294) and National Cancer Institute (CA18029). S.S. is a recipient of a fellowship from the Joel Meyers Memorial Fund.
Authorship
Contribution: S.S. compiled data and wrote the initial manuscript; C.R., Y.-J.K., and T.F. identified the cases; C.R., J.M.K., M.-L.H., C.E.F., and R.H.S. performed PCR and galactomannan assays; C.Y.C., E.S., G.Y., and S.M. performed analysis of NGS; H.X. and T.A.G. performed the statistical analyses; R.C.H. and D.M. prepared and reviewed lung tissue samples; D.N.F., D.K.M., and K.R.J. contributed to the analysis plan; M.B. designed the study and wrote the manuscript; and all authors critically reviewed the manuscript drafts and approved the final version.
Conflict-of-interest disclosure: C.Y.C. is the director of the University of California, San Francisco Abbott Viral Diagnostics and Discovery Center and received research support from Abbott Laboratories Inc. Y.-J.K. received research support from Sanofi Pasteur, Merck, SK Chemicals, Gilead Sciences, and MedImmune. M.B. received research support from Astellas, Chimerix Inc., Genentech/Roche, Merck, Gilead Sciences, and Ansun Biopharma and received consultant fees from Astellas, Chimerix Inc., Genentech/Roche, Merck, Janssen, Clinigen, and Gilead Sciences. The remaining authors declare no competing financial interests.
Correspondence: Michael Boeckh, Vaccine and Infectious Disease Division, Fred Hutchinson Cancer Research Center, 1100 Fairview Ave North, Seattle, WA 98109; e-mail: mboeckh@fredhutch.org.

![Figure 2. Probability of overall survival and respiratory death by presence of pathogens. (A) Kaplan-Meier estimate of overall survival by presence of pathogens. One patient with ciHHV-6 was included in the IPS cases without pathogens. (B) Cumulative incidence of respiratory death by presence of pathogens. (C) Kaplan-Meier estimate of overall survival by pathogen type (aHR for established vs uncertain, 0.83 [0.41-1.69]; P = .61). (D) Cumulative incidence of respiratory death by pathogen type (aHR for established vs uncertain, 0.73 [0.33-1.61]; P = .44).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/24/10.1182_blood-2014-12-617035/4/m_3789f2.jpeg?Expires=1770995978&Signature=HEVJAkzBL6BROMb4uIeIG5FDRaEwtuJCMu-pH0QZrtvUPFpDRF4C4OvYmVepXxwAAByybCYY17qOt-WkdEGgIH4NkhB0mKYMya86Pqd2yveor4AE2bE6ydQJrRqmb7taIm9UALhqd3uIl3YhLRytyOUknznCOSoFxmzHo0v4oV7igh1IedsABP5TeVCLiVeMD-A98X6E~Q5xCZpepBVL1Ac~eDK2khZDLkEyk0mPiv2PP0exQvjDQRqEDbkPMrGB5~DCgNtB74dj6rNaGH~o-o3HvJps0LzmfmbmEp1bmY3jI27A69iy8SDMI~lYiy0HR2IgW0OgFJk25LargVP--w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal