Key Points
Elevated Jun signaling promotes lymphoma growth and dissemination to extranodal sites.
Jun-regulated genes mediate the interaction of malignant cells with stromal cells and adhesion to extracellular matrix proteins.
Abstract
Diffuse large B-cell lymphoma (DLBCL) is an aggressive disease with a high proliferation rate. However, the molecular and genetic features that drive the aggressive clinical behavior of DLBCL are not fully defined. Here, we have demonstrated that activated Jun signaling is a frequent event in DLBCL that promotes dissemination of malignant cells. Downregulation of Jun dramatically reduces lymphoma cell adhesion to extracellular matrix proteins, subcutaneous tumor size in nude mice, and invasive behavior, including bone marrow infiltration and interaction with bone marrow stromal cells. Furthermore, using a combination of RNA interference and gene expression profiling, we identified Jun target genes that are associated with disseminated lymphoma. Among them, ITGAV, FoxC1, and CX3CR1 are significantly enriched in patients with 2 or more extranodal sites. Our results point to activated Jun signaling as a major driver of the aggressive phenotype of DLBCL.
Introduction
The molecular and genetic features that drive the aggressive behavior of B-cell lymphomas are not fully defined. Diffuse large B-cell lymphoma (DLBCL) is an aggressive type of non-Hodgkin lymphoma and the most common type. Gene expression profiling studies have identified at least 3 molecular subtypes of DLBCL that differ in their expression of hundreds of genes and have distinct prognoses.1 The activated B-cell–like subtype (ABC-DLBCL) has the lowest cure rate and is associated with constitutive activation of the nuclear factor κB (NF-κB) pathway.2,3 NF-κB activity requires the CBM signaling complex composed of CARMA1, Bcl10, and MALT1 proteins, and depletion of any of these components is lethal for lymphoma cells.4 Recent studies have also demonstrated that the adaptor protein CARD11 is mutated in 10% to 16% of ABC-DLBCL patient specimens and about 3% of specimens of the germinal center subtype of DLBCL.5-7 Interestingly, in addition to controlling NF-κB activity, CARD11-Bcl10-MALT1 regulates the activator protein 1 (AP-1) transcription factor.8,9
AP-1 is a dimer created by members of the Jun family (c-Jun, JunB, and JunD) with Fos and activating transcription factor family proteins.10 c-Jun and JunB are the best characterized members of the AP-1 family and are frequently overexpressed in several cancers, including lymphoma.11-13 In resting cells, the c-Jun NH2-terminal kinase 2 (JNK2) is complexed with c-Jun and JunB, targeting them for K48-linked polyubiquitination and degradation.14-16 Upon stimulation, Jun proteins are activated and stabilized.16,17 Interestingly, c-Jun is further regulated at the transcriptional level by its own gene product through a positive feedback loop.18 Although it has been demonstrated that Jun transcription factors are primarily involved in regulating the cell cycle under basal conditions,19 a large number of inducible genes contain AP-1–binding sites in their promoters. Therefore, it is not clear how elevated levels of activated Jun affect cell function. In our study, we demonstrated that the c-Jun transcription factor is frequently activated in DLBCL and results in the upregulation of a large number of genes, including those encoding cytokines, surface receptors, and adhesion molecules. Knockdown of Jun dramatically reduces lymphoma cell adhesion to extracellular matrix (ECM) proteins, the size of subcutaneous tumors in nude mice, and invasive behavior such as bone marrow infiltration and interaction with bone marrow stromal cells. Thus, our data indicate that Jun signaling promotes DLBCL growth and dissemination by regulating genes that mediate lymphoma interaction with the microenvironment. Consistent with this conclusion, we found a correlation between c-Jun expression and lymphoma spread to extranodal sites in DLBCL patients.
Methods
Reagents and cell cultures
Antibodies specific for phospho (p)-c-Jun, p-IκBα, JNK2, c-Jun, and CARD11 were purchased from Cell Signaling. Antibodies against JunB, JunD, ubiquitin, and actin were obtained from Santa Cruz. A JNK inhibitor (SP600125) was bought from Sigma. The peptides containing sequences GRGDS and GRGESP were purchased from AnaSpec. CARD11 expression vectors were described previously.9 Short hairpin RNA (shRNA) lentiviral plasmids (pLKO.1-puro) were purchased from Sigma (details available in supplemental Methods, available online on the Blood Web site).
Splenic B cells were purified by using an EasySep mouse B-cell enrichment kit (StemCell Technologies). Bone marrow stromal cells were prepared from femurs and tibias of SCID mice, and adherent cells were enriched by culture in RPMI 1640 medium with 10% fetal bovine serum (FBS) for 5 days. Human DLBCL cell lines were cultured in Iscove modified Dulbecco medium supplemented with 2-mercaptoethanol, antibiotics, and 15% FBS (OCI-Ly3 and OCI-Ly7) or 15% to 20% human plasma (OCI-Ly10). Cells were grown in 5% CO2 at 37°C and passed every 3 days. M2-10B4 bone marrow fibroblasts were cultured in RPMI 1640 plus 10% FBS and antibiotics. HS-5 cells were maintained in Dulbecco’s modified Eagle medium supplemented with 10% FBS and antibiotics. Stable transfection of OCI-Ly7 cells was established by 2 rounds of lentiviral infection. The transfection efficiency was determined by flow cytometry and western blot analysis.
Xenograft model of DLBCL
Nude mice were maintained under pathogen-free conditions in the institutional animal facility. For xenograft studies, mice were inoculated with 5 × 106 OCI-Ly3 cells expressing different shRNAs. The cells were mixed with Matrigel (BD Biosciences) in a 1:1 ratio. Tumor volume was determined by weekly digital caliper measurements and calculated by using the formula (length × width squared)/2. All experiments were performed in compliance with the institutional guidelines and according to the protocol approved by the Institutional Animal Care and Use Committee.
Immunoblotting and immunoprecipitation
Cells were lysed in a buffer containing 1% nonidet P-40 as described before.8 Total cell lysates were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and western blotting or immunoprecipitated with c-Jun antibody. The immunoprecipitates were washed with radioimmunoprecipitation assay buffer, eluted with 2× SDS loading buffer, and fractionated on 10% SDS-PAGE. Immunoblots were incubated with specific primary antibodies and then with horseradish peroxidase (HRP) –conjugated secondary antibodies and developed according to the manufacturer’s protocol (Pierce).
Cytokine array
The array for human cytokine detection was purchased from Millipore (Milliplex Map Human Cytokine/Chemokine magnetic bead panel immunoassay, #HCYTMAG-60K-PX38). The assay was performed according to the manufacturer’s protocols, and the mean fluorescence intensities were detected with a Luminex 200 system. The data were collected by using xPONENT software and analyzed by using Prism GraphPad software. Results are presented as the arithmetic mean of triplicates.
Quantitative polymerase chain reaction
Total RNA was isolated by using an RNeasy kit (QIAGEN) and reverse transcribed by using SuperScriptIII (Invitrogen). Quantitative polymerase chain reaction (qPCR) was performed with Power SYBR Green PCR Master Mix (Applied Biosystems). The amount of transcript was normalized to glyceraldehyde-3-phosphate dehydrogenase. Melt curves were run to ensure amplification of a single product.
Electrophoretic mobility shift assay
Extraction of nuclear proteins was performed as described before,8 and protein concentration was determined by the Bio-Rad protein assay using bovine serum albumin as the standard. Nuclear extracts (4 μg) were incubated with a 32P-labeled, double-stranded AP-1 or Oct1-specific oligonucleotide probe (Promega) for 15 minutes at room temperature. For the supershift, nuclear extracts were incubated with antibody specific to AP-1 members before the probe was added. Samples were fractionated on a 5% polyacrylamide gel and visualized by autoradiography.
Cell proliferation and adhesion
Cell proliferation was determined by using the 3-(4,5-dimethylthiazol-2-yl)-2,5-dimethyltetrazolium bromide assay. Adhesion of cells to ECM proteins was determined by using an ECM Cell Adhesion Array Kit (Millipore, #ECM540). The experiments were performed in triplicate, and the results are presented as means + standard deviations.
Tumor specimens
The primary tumor specimens (archival) were provided by the Hematopathology Tissue Bank of MD Anderson Cancer Center. Immunohistochemical stains were performed on formalin-fixed paraffin-embedded tissues. Antibody specific for p-c-Jun (Ser73) was purchased from Cell Signaling (#9164).
Gene expression profiling
Microarray experiments with DLBCL cell lines were performed by using the Agilent platform (Agilent Whole Human Genome Microarray, 4 × 44 k) and analyzed by using GeneSpringGX12 software. Signals from control cells (Cy3) were compared with signals from the respective modified cells (Cy5). For clinical samples, gene expression data were retrieved from a public repository (GSE10846). Log2-transformed and median-centered intensities were used to compare c-Jun, FoxC1, ITGAV, and CX3CR1 expression levels. The Spearman test was used to analyze the correlation between c-Jun and selected genes in tumor tissue samples. A P value < .05 was considered to indicate a significant difference.
Results
Level of AP-1 transcription factors is elevated in DLBCL
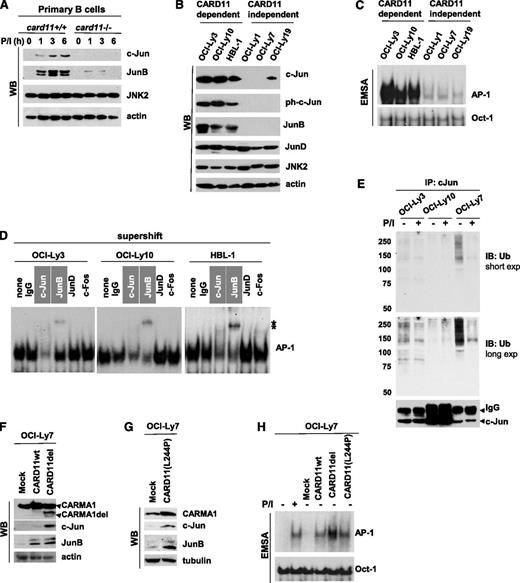
Our previous studies had indicated that antigen-receptor stimulation results in CARD11-dependent JNK2 activation and enhances expression of the AP-1 transcription factors c-Jun and JunB in T cells.8,9 However, the role of CARD11 in Jun signaling in B cells has not been defined. Therefore, we isolated splenic B cells from card11+/+ and card11−/− mice and noted a signal-dependent induction of c-Jun and JunB. Stimulation of wild-type (wt) cells with the mitogen phorbol myristate acetate resulted in the accumulation of c-Jun and JunB, whereas expression of these proteins was impaired in the absence of CARD11 (Figure 1A). On the basis of this finding, we hypothesized that elevated CARD11 activity leads to the accumulation of activated Jun transcription factors in B cells.
CARD11 regulates c-Jun and JunB level in primary B cells and DLBCL cells. (A) Murine primary B cells were isolated from spleens of wt or CARMA1-deficient mice. The cells (4 × 106 cells per sample) were stimulated with phorbol myristate acetate (PMA) plus ionomycin (P/I; 20 and 100 ng/mL, respectively) for various time points. Whole-cell lysates were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by western blotting using antibodies against c-Jun, JunB, JNK2, or β-actin. (B) Cell lysates were prepared from the indicated lymphoma cell lines and subjected to SDS-PAGE followed by western blotting. (C) Nuclear extracts were prepared from DLBCL cells and analyzed by electrophoretic mobility shift assay (EMSA) by using 32P-labeled probes containing AP-1– or Oct-1–binding sites. (D) For the supershift assay, nuclear extracts were pre-incubated (30 minutes at 4°C) with the indicated antibodies and then subjected to EMSA. (E) c-Jun ubiquitination in lymphoma cell lines. The cells were stimulated with P/I for 5 minutes, and the cell lysates were precipitated with c-Jun antibodies for 16 hours. The immunocomplexes (top panel) and whole-cell lysates (bottom panel) were subjected to SDS-PAGE and analyzed by western blotting by using ubiquitin or c-Jun antibodies. (F-G) OCI-Ly7 cells were stably transfected withCARD11wt, constitutively active CARD11del (the linker region between the coiled-coil and PDZ domains of CARD11 was deleted) or CARD11(L244P) mutant by lentiviral infection. Whole-cell lysates were subjected to SDS-PAGE followed by western blot analysis using the indicated antibodies. (H) OCI-Ly7 cells or cells transduced with the indicated CARD11 constructs were subjected to EMSA to determine AP-1 binding activity. Exp, exposure; IB, immunoblotting; IgG, heavy chain; IP, immunoprecipitation; ph, phosphorylated; Ub, ubiquitin; WB, western blotting; *, shifted band.
CARD11 regulates c-Jun and JunB level in primary B cells and DLBCL cells. (A) Murine primary B cells were isolated from spleens of wt or CARMA1-deficient mice. The cells (4 × 106 cells per sample) were stimulated with phorbol myristate acetate (PMA) plus ionomycin (P/I; 20 and 100 ng/mL, respectively) for various time points. Whole-cell lysates were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by western blotting using antibodies against c-Jun, JunB, JNK2, or β-actin. (B) Cell lysates were prepared from the indicated lymphoma cell lines and subjected to SDS-PAGE followed by western blotting. (C) Nuclear extracts were prepared from DLBCL cells and analyzed by electrophoretic mobility shift assay (EMSA) by using 32P-labeled probes containing AP-1– or Oct-1–binding sites. (D) For the supershift assay, nuclear extracts were pre-incubated (30 minutes at 4°C) with the indicated antibodies and then subjected to EMSA. (E) c-Jun ubiquitination in lymphoma cell lines. The cells were stimulated with P/I for 5 minutes, and the cell lysates were precipitated with c-Jun antibodies for 16 hours. The immunocomplexes (top panel) and whole-cell lysates (bottom panel) were subjected to SDS-PAGE and analyzed by western blotting by using ubiquitin or c-Jun antibodies. (F-G) OCI-Ly7 cells were stably transfected withCARD11wt, constitutively active CARD11del (the linker region between the coiled-coil and PDZ domains of CARD11 was deleted) or CARD11(L244P) mutant by lentiviral infection. Whole-cell lysates were subjected to SDS-PAGE followed by western blot analysis using the indicated antibodies. (H) OCI-Ly7 cells or cells transduced with the indicated CARD11 constructs were subjected to EMSA to determine AP-1 binding activity. Exp, exposure; IB, immunoblotting; IgG, heavy chain; IP, immunoprecipitation; ph, phosphorylated; Ub, ubiquitin; WB, western blotting; *, shifted band.
Because CARD11 is frequently deregulated in DLBCL cells, we speculated that these cells have elevated Jun protein levels. To test this idea, we used cell lines that had been shown to depend on CARD11 to survive (OCI-Ly3, OCI-Ly10, and HBL1) as well as CARD11-independent cell lines (OCI-Ly1, OCI-Ly7, and OCI-Ly19).4,5 Of note, CARD11 is mutated in OCI-Ly3, but OCI-Ly10 and HBL1 cells carry oncogenic mutations upstream of CARD11 that result in chronic activation of this protein. We found that the amount of c-Jun and JunB was much higher in CARD11-dependent cells (Figure 1B). Interestingly, the level of JunD, the third member of the Jun family, was comparable in all tested cell lines. Consistent with our results from the immunoblotting experiment, we found that AP-1 was selectively activated in CARD11-dependent DLBCL cells (Figure 1C). Supershift analysis further revealed that active AP-1 dimers were composed of c-Jun and JunB in these cells (Figure 1D).
It has been shown that c-Jun is constitutively ubiquitinated and degraded in resting cells16 but that c-Jun phosphorylation by JNK leads to protein stabilization.17 To examine the effect of CARD11 activity on c-Jun ubiquitination, we immunoprecipitated c-Jun from unstimulated cells and cells treated with phorbol myristate acetate plus ionomycin (Figure 1E). We found lower levels of c-Jun ubiquitination in both OCI-Ly3 and OCI-Ly10 cells than in OCI-Ly7 cells. Moreover, ubiquitination of c-Jun was strongly suppressed upon stimulation only in OCI-Ly7 cells (Figure 1E), suggesting that c-Jun is more stable in the presence of activated CARD11. These results are consistent with our previously published data that c-Jun is stabilized upon antigen-receptor stimulation in T cells.9
To further demonstrate that accumulation of c-Jun and JunB is mediated by activated CARD11, OCI-Ly7 cells were stably transfected with the previously described CARD11 mutant (CARD11del)9,20 or CARD11 cloned from OCI-Ly3 cells (L244P-activating mutation). Indeed, both CARD11 mutants strongly increased the amount of c-Jun and JunB compared with that of mock (Figure 1F-G) and CARD11wt-expressing controls (Figure 1F). CARD11del and CARD11(L244P) consistently induced AP-1 activity in transduced cells, and their effect was stronger than that of CARD11wt (Figure 1G). Together, our data indicate that CARD11 mediates elevated Jun signaling in DLBCL.
Cell aggregation and adhesion is reduced by c-Jun knockdown
Previous studies demonstrated that Jun transcription factors play important roles in cell proliferation19 ; thus, elevated CARMA1/JNK2/Jun signaling might result in enhanced cell proliferation. Because both c-Jun and JunB are regulated by CARD119 and these factors have overlapping as well as distinct functions,19 we decided to knock down both proteins in OCI-Ly3 and OCI-Ly10 cells (Figure 2A). Five different c-Jun- and JunB-specific shRNAs were tested (supplemental Figure 1A-C), and cells with the highest knockdown efficiency were selected for functional assays. Although cells with reduced amounts of both Jun factors proliferated significantly slower in in vitro culture, the inhibitory effect seemed to be only partial (Figure 2B). Similar results were obtained by using SP600125, a pharmacological JNK inhibitor that only slightly decreased the cell proliferation rate and increased the number of apoptotic cells (supplemental Figure 2). Intriguingly, we observed morphologic changes and reduced aggregation of cells that were transduced with c-Jun/JunB shRNAs (Figure 2C and supplemental Figure 1D). Because the CD44 adhesion molecule is a known c-Jun target that mediates cell-cell interaction, we confirmed that silencing of c-Jun correlated with reduced expression of CD44 (supplemental Figure 1). Conversely, transduction of the Jun-negative cell line OCI-Ly7 with the CARD11 mutant resulted in spontaneous cell aggregation and induction of CD44 (supplemental Figure 3).
Cellular adhesion is reduced upon c-Jun knockdown. (A) Efficiency of c-Jun and JunB knockdown in ABC-DLBCL cell lines using specific shRNAs. Cell lysates were prepared from the indicated cell lines and subjected to SDS-PAGE followed by western blotting using the indicated antibodies. The densitometric analysis was performed by using ImageJ software. (B) Proliferation assay. Cells were cultured in 96-well plates and 3-(4,5-dimethylthiazol-2-yl)-2,5-dimethyltetrazolium bromide (MTT) was added to the cells 4 hours before lysis in isopropanol with 1% hydrochloric acid. The plate was read with a microplate spectrometer using a 570-nm filter. Results are presented as the mean + standard deviation (SD) of triplicate cultures. Statistical significance was evaluated by the Student t test. (C) Loss of cellular aggregation and adhesion upon Jun knockdown. OCI-Ly10 cells were cultured in Iscove modified Dulbecco medium (IMDM) medium with different concentrations of human plasma for 24 hours. The images were taken with the Olympus IX70 inverted fluorescence microscope (×20). (D) The cells attached to the bottom of the culture dish (16 hours) were quantified by using colorimetric detection. Results are shown as the mean + SD of triplicate cultures. (E) Adhesion of lymphoma cell lines to the indicated ECM proteins (4 hours) was determined by using the ECM Cell Adhesion Array Kit (Millipore). Results are shown as the mean ± SD of triplicate cultures. (F) Cell treatment with peptides containing the RGD or RGE sequence (30 minutes) was followed with the cell adhesion assay. OD, optical density; FNK, fibronectin; RGD, Arg-Gly-Asp motif.
Cellular adhesion is reduced upon c-Jun knockdown. (A) Efficiency of c-Jun and JunB knockdown in ABC-DLBCL cell lines using specific shRNAs. Cell lysates were prepared from the indicated cell lines and subjected to SDS-PAGE followed by western blotting using the indicated antibodies. The densitometric analysis was performed by using ImageJ software. (B) Proliferation assay. Cells were cultured in 96-well plates and 3-(4,5-dimethylthiazol-2-yl)-2,5-dimethyltetrazolium bromide (MTT) was added to the cells 4 hours before lysis in isopropanol with 1% hydrochloric acid. The plate was read with a microplate spectrometer using a 570-nm filter. Results are presented as the mean + standard deviation (SD) of triplicate cultures. Statistical significance was evaluated by the Student t test. (C) Loss of cellular aggregation and adhesion upon Jun knockdown. OCI-Ly10 cells were cultured in Iscove modified Dulbecco medium (IMDM) medium with different concentrations of human plasma for 24 hours. The images were taken with the Olympus IX70 inverted fluorescence microscope (×20). (D) The cells attached to the bottom of the culture dish (16 hours) were quantified by using colorimetric detection. Results are shown as the mean + SD of triplicate cultures. (E) Adhesion of lymphoma cell lines to the indicated ECM proteins (4 hours) was determined by using the ECM Cell Adhesion Array Kit (Millipore). Results are shown as the mean ± SD of triplicate cultures. (F) Cell treatment with peptides containing the RGD or RGE sequence (30 minutes) was followed with the cell adhesion assay. OD, optical density; FNK, fibronectin; RGD, Arg-Gly-Asp motif.
Intriguingly, we have also observed Jun-dependent cell attachment to the bottom of the culture dish in the presence of a high concentration of human plasma (Figure 2C-D). This effect might be mediated by ECM proteins; therefore, we determined the role of Jun in lymphoma cell interaction with a panel of ECM proteins. Both OCI-Ly3 (Figure 2E, upper graph) and OCI-Ly10 cells (Figure 2E, lower graph) attached to the plate coated with multiple ECM proteins (such as fibronectin, vitronectin, collagen IV, and tenascin) in the serum-free conditions, whereas this attachment was lost upon c-Jun and JunB knockdown. Our results suggest that the observed phenomenon might be associated with the expression of integrins, such as integrin αvβ3 or αvβ5, that bind to fibronectin and vitronectin through the Arg-Gly-Asp (RGD) motif.21 To test this possibility, we pre-incubated lymphoma cells with the synthetic RGD peptide (or ArgGlyGlu [RGE] as a negative control) and then determined lymphoma cell adhesion to fibronectin-coated plates (Figure 2F). Indeed, this treatment abolished cell adhesion ability. Together, our data indicate that the genes regulated by the Jun family of transcription factors contribute to cell-cell interaction and the adhesion of DLBCL cells to ECM.
c-Jun and JunB promote tumor growth in a xenograft model of DLBCL
To determine the role of elevated Jun signaling in vivo, we used a xenograft model of DLBCL. In contrast to the results from in vitro studies, we observed a dramatic reduction in tumor volume and weight after c-Jun and JunB knockdown (Figure 3A), even when the mice were injected with both control and modified OCI-Ly3 cells (bilateral implantation). Furthermore, gross examination of the experimental mice revealed enlarged lymph nodes (Figure 3B, left panel), which suggested lymphoma cell dissemination from the primary site. We did detect infiltration of human CD20+ cells in peripheral lymph nodes and spleens (Figure 3B, right panel). Similarly, when mice were implanted with a single cell line (one-site implantation), we found that tumor volume and weight were significantly reduced (P < .014) upon JunB and c-Jun knockdown (Figure 3C). Although we observed dissemination of lymphoma cells into lymph nodes in both groups of mice (data not shown), the involvement of bone marrow was reduced in mice that had received cells with downregulated Jun (Figure 3D). To rule out the possibility that the difference in bone marrow involvement was affected by the size of the primary tumor, we analyzed only those mice that generated subcutaneous tumors of similar sizes (Figure 3D, right panel, marked as #1, #2, and #3).
Knockdown of c-Jun and JunB reduces tumor size in a xenograft model of DLBCL and lymphoma cell invasiveness. (A) Xenograft model of DLBCL. Nude mice (n = 5) were inoculated with OCI-Ly3 cells (5 × 106) stably expressing shGFP (right site) and shJunB/c-Jun (left site). Tumors were measured weekly with calipers, and tumor weight was determined at week 7. (B) Enlarged lymph nodes in nude mice inoculated subcutaneously with OCI-Ly3 cells expressing the indicated shRNAs. Lymphoma cells were identified by immunostaining (IHC) with human CD20 antibody (Ab; brown). Pictures were taken with the Olympus IX70 inverted fluorescence microscope (×40). (C) Subcutaneous tumors in nude mice. Control cells or cells with silenced JunB/c-Jun were injected into experimental mice (n = 8 per group), and tumor growth was monitored as described in (A). (D) Bones (femurs and tibias) were collected from experimental mice, and bone marrow infiltration by CD20+ cells was quantified by using the Automated Cellular Imaging System (ACIS III, Dako). Results are presented as relative intensity level in mice that developed tumors of comparable size (labeled as #1, #2, and #3). (E) In vitro invasion assay was performed by using transwell inserts coated with Matrigel. Results are presented as mean + SD of triplicate cultures. (F) Lymphoma cell lines were labeled with CMFDA (CellTracker Green, Molecular Probes) for 30 minutes and injected into the recipient mice (106 cells per mouse). Bone marrow infiltration was determined by flow cytometry 16 hours later. Results are presented as mean + SD of duplicates. (G) Lymphoma cell adhesion to bone marrow stroma. Freshly isolated bone marrow stromal cells were seeded in 6-well plates. OCI-Ly3 cells stably expressing shGFP (control) or shJunB plus c-Jun were labeled with 5-chloromethylfluorescein diacetate (CellTracker Green) for 30 minutes and cocultured with a monolayer of stromal cells for 3 hours. The pictures were taken after 3 washes with warm PBS.
Knockdown of c-Jun and JunB reduces tumor size in a xenograft model of DLBCL and lymphoma cell invasiveness. (A) Xenograft model of DLBCL. Nude mice (n = 5) were inoculated with OCI-Ly3 cells (5 × 106) stably expressing shGFP (right site) and shJunB/c-Jun (left site). Tumors were measured weekly with calipers, and tumor weight was determined at week 7. (B) Enlarged lymph nodes in nude mice inoculated subcutaneously with OCI-Ly3 cells expressing the indicated shRNAs. Lymphoma cells were identified by immunostaining (IHC) with human CD20 antibody (Ab; brown). Pictures were taken with the Olympus IX70 inverted fluorescence microscope (×40). (C) Subcutaneous tumors in nude mice. Control cells or cells with silenced JunB/c-Jun were injected into experimental mice (n = 8 per group), and tumor growth was monitored as described in (A). (D) Bones (femurs and tibias) were collected from experimental mice, and bone marrow infiltration by CD20+ cells was quantified by using the Automated Cellular Imaging System (ACIS III, Dako). Results are presented as relative intensity level in mice that developed tumors of comparable size (labeled as #1, #2, and #3). (E) In vitro invasion assay was performed by using transwell inserts coated with Matrigel. Results are presented as mean + SD of triplicate cultures. (F) Lymphoma cell lines were labeled with CMFDA (CellTracker Green, Molecular Probes) for 30 minutes and injected into the recipient mice (106 cells per mouse). Bone marrow infiltration was determined by flow cytometry 16 hours later. Results are presented as mean + SD of duplicates. (G) Lymphoma cell adhesion to bone marrow stroma. Freshly isolated bone marrow stromal cells were seeded in 6-well plates. OCI-Ly3 cells stably expressing shGFP (control) or shJunB plus c-Jun were labeled with 5-chloromethylfluorescein diacetate (CellTracker Green) for 30 minutes and cocultured with a monolayer of stromal cells for 3 hours. The pictures were taken after 3 washes with warm PBS.
Together, our results suggest that elevated Jun signaling contributes to the aggressive phenotype of lymphoma cells and supports lymphoma growth in the bone marrow microenvironment. To test these possibilities, we performed an in vitro invasion assay and found that downregulation of Jun proteins dramatically suppressed cell invasion ability in Matrigel (Figure 3E). Next, we assessed bone marrow infiltration after tail-vein injection. Consistent with previous data, we detected significantly fewer lymphoma cells in bone marrow isolated from mice that received cells with c-Jun and JunB knockdown (Figure 3F). Finally, to determine whether silencing of Jun affects the interaction of lymphoma cells with bone marrow stroma, we cocultured OCI-Ly3 with primary bone marrow stromal cells (Figure 3G) or cells of the previously established M2-10B4 stromal cell line (supplemental Figure 4). We found that control lymphoma cells strongly adhered to the monolayer of stromal cells during 3 hours of coculture; however, depletion of Jun proteins led to greatly reduced adhesion of lymphoma cells to the bone marrow-derived stromal cells. Together, our data suggest that Jun signaling promotes lymphoma spread to extranodal sites such as bone marrow and interaction with stromal cells.
Jun expression is increased in patients with multiple extranodal involvement
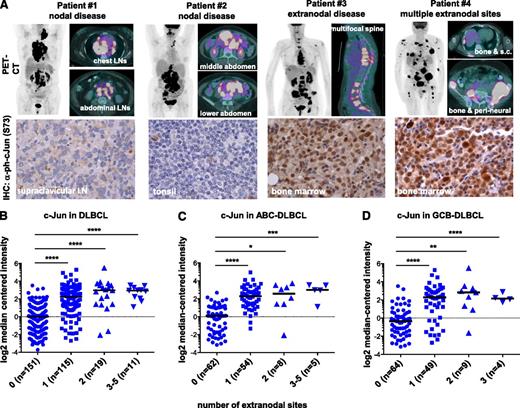
Elevated Jun signaling might promote the dissemination of DLBCL, especially to extranodal sites. To test this hypothesis, we performed immunohistochemical analysis of tumor biopsy specimens collected from 25 patients with advanced disease (stage III or IV including 9 bone marrow–positive cases) (Table 1). Although nuclear expression of p-c-Jun varied among the examined specimens, we found strong staining in all bone marrow–positive cases. Furthermore, nuclear p-c-Jun was detected in patients with extranodal disease identified on positron emission tomography/computed tomography scans, and representative images are shown in Figure 4A. Next, we used the microarray technique to compare the c-Jun expression levels in tumor specimens obtained from patients with nodal disease with those from extranodal cases, including disseminated lymphoma (Figure 4B). Complementary DNA microarray data and clinical information for 414 patients (296 with a known number of extranodal sites) were retrieved from a public repository (GSE10846) and analyzed by using Oncomine software. High c-Jun expression was associated mainly with extranodal localization of lymphoma lesions (P < .0001) (Figure 4B and supplemental Figure 5). The same trend was observed when we grouped patients on the basis of the ABC and germinal center B-cell (GCB) subtypes of DLBCL (Figure 4C-D), suggesting that elevated Jun signaling is not restricted to ABC-DLBCL. Indeed, by analyzing the established ABC- and GCB-DLBCL cell lines, we confirmed that elevated Jun is more common in but not unique to the ABC subtype (supplemental Figure 6). Together, our findings suggest that c-Jun promotes lymphoma dissemination to extranodal sites and that this effect is independent on ABC and/or GCB classification.
Immunohistochemical analysis of p-c-Jun (Ser73) expression in a subset of 25 DLBCL tumor tissues collected from patients with or without bone marrow involvement
| Nuclear p-c-Jun (Ser73) staining . | Nodal disease (n = 4) . | Extranodal disease . | ||||
|---|---|---|---|---|---|---|
| Without bone marrow involvement (n = 12) . | With bone marrow involvement (n = 9) . | |||||
| No. . | % . | No. . | % . | No. . | % . | |
| Negative | 3 | 75 | 2 | 17 | 0 | |
| Weak to moderate | 1 | 25 | 7 | 58 | 0 | |
| Strong | 0 | 3 | 25 | 9 | 100 | |
| Nuclear p-c-Jun (Ser73) staining . | Nodal disease (n = 4) . | Extranodal disease . | ||||
|---|---|---|---|---|---|---|
| Without bone marrow involvement (n = 12) . | With bone marrow involvement (n = 9) . | |||||
| No. . | % . | No. . | % . | No. . | % . | |
| Negative | 3 | 75 | 2 | 17 | 0 | |
| Weak to moderate | 1 | 25 | 7 | 58 | 0 | |
| Strong | 0 | 3 | 25 | 9 | 100 | |
c-Jun is enriched in extranodal lymphoma. (A) Positron emission tomography/computed tomography (PET/CT) images from patients with different lymphoma localization. Patients #1 and #2 had only nodal sites of involvement; representative images are shown. Patient #3 had multifocal extranodal involvement in the bone marrow in the spine, as shown in sagittal images. Patient #4 had multiple extranodal sites of involvement, including bone marrow, subcutaneous tissues, and perineural spread. Expression of p-c-Jun (Ser73) in DLBCL tumor tissues from these patients was determined by immunohistochemistry; representative images are shown (×500). (B-D) Expression of c-Jun was stratified by the number of extranodal sites using microarray data available in a public repository (GSE10846). Log2 median-centered intensities of c-Jun (probe 201465_s) in 296 cases with a known number of extranodal sites were obtained by using Oncomine v4.5 (www.oncomine.org). Patients diagnosed with the ABC or GCB subtypes of DLBCL were analyzed in (C) and (D), respectively. Statistical significance was evaluated by using two-tailed Student t test. ****P < .0001; ***P < .001; **P < .01; *P < .05.
c-Jun is enriched in extranodal lymphoma. (A) Positron emission tomography/computed tomography (PET/CT) images from patients with different lymphoma localization. Patients #1 and #2 had only nodal sites of involvement; representative images are shown. Patient #3 had multifocal extranodal involvement in the bone marrow in the spine, as shown in sagittal images. Patient #4 had multiple extranodal sites of involvement, including bone marrow, subcutaneous tissues, and perineural spread. Expression of p-c-Jun (Ser73) in DLBCL tumor tissues from these patients was determined by immunohistochemistry; representative images are shown (×500). (B-D) Expression of c-Jun was stratified by the number of extranodal sites using microarray data available in a public repository (GSE10846). Log2 median-centered intensities of c-Jun (probe 201465_s) in 296 cases with a known number of extranodal sites were obtained by using Oncomine v4.5 (www.oncomine.org). Patients diagnosed with the ABC or GCB subtypes of DLBCL were analyzed in (C) and (D), respectively. Statistical significance was evaluated by using two-tailed Student t test. ****P < .0001; ***P < .001; **P < .01; *P < .05.
Jun signaling mediates cytokine production by DLBCL
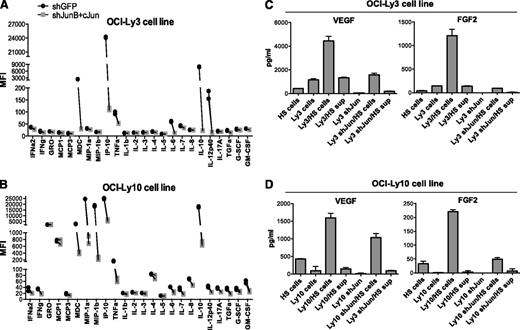
Interaction of lymphoma cells with the microenvironment involves the production of various chemokines, such as interleukin-6 (IL-6) and IL-10, that promote the proliferation and survival of B cells.22,23 Previous studies and our own data (supplemental Figure 7A) have indicated that OCI-Ly3 and OCI-Ly10 cells spontaneously release IL-6 and IL-10.24 To further determine the role of Jun transcription factors in cytokine/chemokine production by OCI-Ly3 and OCI-Ly10 cells, we used the multiplex suspension array system. We found that knockdown of c-Jun and JunB effectively blocked the release of macrophage-derived chemokine, interferon-inducible protein 10, IL-6, IL-10, and IL-12 (Figure 5A-B). Of note, these cytokines were not secreted by Jun-negative DLBCL cell lines (supplemental Figure 7B).
Downregulation of c-Jun reduces cytokine production by lymphoma cells. (A-B) Multiplexed cytokine array was used to detect cytokines released by OCI-Ly3 and OCI-Ly10 cells during 48 hours of culture in complete medium. Results are shown as the mean fluorescence intensity (MFI) determined in duplicates from 2 independent experiments. (C-D) OCI-Ly3 or OCI-Ly10 cells were cocultured with HS-5 mesenchymal cells or supernatants (sup) from HS-5 cells (30%) for 48 hours. Concentrations of VEGF and FGF2 were measured by using a cytokine multiplex. Results are presented as mean + SD of duplicates from 2 independent experiments. FGF2, basic fibroblast growth factor; G-CSF, granulocyte colony-stimulating factor; GM-CSF, granulocyte-macrophage colony-stimulating factor; GRO, growth-regulated oncogene; IFN, interferon; IP-10, interferon-induced protein 10; MCP, monocyte chemoattractant protein; MDC, macrophage-derived chemokine; MIP, macrophage inflammatory protein; TGF, transforming growth factor; TNF, tumor necrosis factor; VEGF, vascular endothelial growth factor.
Downregulation of c-Jun reduces cytokine production by lymphoma cells. (A-B) Multiplexed cytokine array was used to detect cytokines released by OCI-Ly3 and OCI-Ly10 cells during 48 hours of culture in complete medium. Results are shown as the mean fluorescence intensity (MFI) determined in duplicates from 2 independent experiments. (C-D) OCI-Ly3 or OCI-Ly10 cells were cocultured with HS-5 mesenchymal cells or supernatants (sup) from HS-5 cells (30%) for 48 hours. Concentrations of VEGF and FGF2 were measured by using a cytokine multiplex. Results are presented as mean + SD of duplicates from 2 independent experiments. FGF2, basic fibroblast growth factor; G-CSF, granulocyte colony-stimulating factor; GM-CSF, granulocyte-macrophage colony-stimulating factor; GRO, growth-regulated oncogene; IFN, interferon; IP-10, interferon-induced protein 10; MCP, monocyte chemoattractant protein; MDC, macrophage-derived chemokine; MIP, macrophage inflammatory protein; TGF, transforming growth factor; TNF, tumor necrosis factor; VEGF, vascular endothelial growth factor.
Next, we examined the requirement of c-Jun in stroma-induced cytokine production. Interestingly, we found that coculture of lymphoma cells with human HS-5 mesenchymal cells, but not supernatants from HS-5 cells, strongly induced or enhanced the secretion of 2 proangiogenic mediators, vascular endothelial growth factor (VEGF) and basic fibroblast growth factor (FGF2) (Figure 5C-D). This effect was reduced when shJun-expressing cells were incubated with an HS-5 monolayer. Thus, Jun transcription factors are important mediators of signals received by lymphoma cells upon interaction with stroma.
Identification of Jun-regulated genes in DLBCL
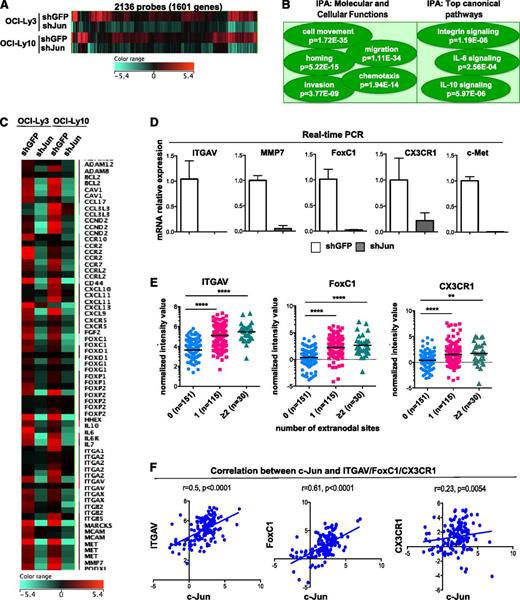
Collectively, our data indicate that constitutively active Jun signaling leads to the upregulation of genes involved in lymphoma cell invasion and interaction with the microenvironment. To identify these genes, we performed complementary DNA microarray analysis and compared the gene expression profiles of OCI-Ly3 and OCI-Ly10 cells transduced with shGFP (control) or Jun-specific shRNAs. We found that silencing of Jun transcription factors significantly suppressed the expression of hundreds of genes (P < .05) (Figure 6A), and analysis with Ingenuity Pathway Analysis tools revealed that affected genes were associated with cellular functions such as cell movement, migration, chemotaxis, homing, and invasion (Figure 6B). Among genes downregulated at least twofold in both cell lines (supplemental Table 1), we identified multiple signaling molecules (FoxP1, FoxC1), adhesion molecules (CD44, ITGAV, ITGB2, ITGB5), surface receptors (Met, CCR5, CCR7, IL-6R), enzymes (matrix metalloproteinase 7), and cytokines and/or chemokines (Figure 6C). These results were further validated by real-time PCR (Figure 6D and supplemental Figure 8).
Effect of Jun knockdown on gene expression in DLBCL cell lines. (A) Genes downregulated in lymphoma cells upon Jun knockdown. Microarray analysis was performed with OCI-Ly3 and OCI-Ly10 cells stably expressing shGFP (control) or shJunB/c-Jun. Heatmap was generated by using GeneSpringGX12 software. (B) Ingenuity Pathway Analysis (IPA) software was used with microarray data obtained in (A). (C) Heatmap of selected genes that were downregulated in lymphoma cell lines upon Jun knockdown. (D) Loss of ITGAV, MMP7, FoxC1, CX3CR1, and Met mRNA expression upon c-Jun/JunB knockdown was quantified by real-time PCR. qPCR was performed in triplicate using Power SYBR Green PCR Master Mix. The amounts of transcript were normalized to glyceraldehyde-3-phosphate dehydrogenase. Melt curves were run to ensure amplification of a single product. Results are presented as mean + SD of triplicates. (E) Expression of ITGAV, FoxC1, and CX3CR1 was stratified by number of extranodal sites. Analysis was performed on arrays available in the public repository (GSE10846). Statistical significance was evaluated by using 2-tailed Student t test. ****P < .0001; **P < .002. (F) Correlation between c-Jun and ITGAV (left), FoxC1 (middle), and CX3CR1 (right) expression in DLBCL cases with secondary extranodal involvement.
Effect of Jun knockdown on gene expression in DLBCL cell lines. (A) Genes downregulated in lymphoma cells upon Jun knockdown. Microarray analysis was performed with OCI-Ly3 and OCI-Ly10 cells stably expressing shGFP (control) or shJunB/c-Jun. Heatmap was generated by using GeneSpringGX12 software. (B) Ingenuity Pathway Analysis (IPA) software was used with microarray data obtained in (A). (C) Heatmap of selected genes that were downregulated in lymphoma cell lines upon Jun knockdown. (D) Loss of ITGAV, MMP7, FoxC1, CX3CR1, and Met mRNA expression upon c-Jun/JunB knockdown was quantified by real-time PCR. qPCR was performed in triplicate using Power SYBR Green PCR Master Mix. The amounts of transcript were normalized to glyceraldehyde-3-phosphate dehydrogenase. Melt curves were run to ensure amplification of a single product. Results are presented as mean + SD of triplicates. (E) Expression of ITGAV, FoxC1, and CX3CR1 was stratified by number of extranodal sites. Analysis was performed on arrays available in the public repository (GSE10846). Statistical significance was evaluated by using 2-tailed Student t test. ****P < .0001; **P < .002. (F) Correlation between c-Jun and ITGAV (left), FoxC1 (middle), and CX3CR1 (right) expression in DLBCL cases with secondary extranodal involvement.
Interestingly, our profiling of DLBCL cell lines expressing Jun-specific shRNAs revealed three novel Jun-regulated genes (vitronectin receptor ITGAV, transcription factor FoxC1, and fractalkine receptor CX3CR1; Figure 6B) that are implicated in tumor migration and invasion.25-27 Real-time PCR confirmed that knockdown of c-Jun and JunB resulted in almost complete depletion of ITGAV, FoxC1, and CX3CR1 in OCI-Ly3 cells (Figure 6D). Therefore, we assessed the expression levels of these genes in DLBCL biopsy samples and found significant enrichment in patients with secondary extranodal localization (Figure 6E). Importantly, there was a positive correlation between c-Jun and ITGAV (r = .5; P < .0001), FoxC1 (r = .61; P < .0001), or CX3CR1 (r = .23; P < .0054) in extranodal tumor samples (Figure 6F). Thus, our results suggest that these genes play an important role in Jun-mediated dissemination of DLBCL.
Discussion
This study highlights the role of CARD11-Jun signaling in DLBCL biology, particularly in lymphoma cell dissemination and interaction with the microenvironment. We demonstrated that elevated CARD11 activity in DLBCL results in the accumulation of c-Jun and JunB and in AP-1 activation in the absence of exogenous stimulus.
CARD11 is a scaffold molecule expressed exclusively in hematopoietic cells.28,29 Its role in antigen-induced NF-κB activation is well established,30-34 and our previous study linked CARD11 to the signal-dependent induction of c-Jun and JunB in T cells.8 Here, we further demonstrated that elevated CARD11 activity drives the activation of c-Jun and JunB in DLBCL. The molecular mechanism by which stimulation of B-cell receptor induces accumulation of activated c-Jun is poorly understood. c-Jun is an unstable protein that constantly undergoes ubiquitination and degradation,16 whereas signal-dependent phosphorylation of c-Jun leads to its stabilization.17 In this study, we found that c-Jun ubiquitination was barely detectable in OCI-Ly3 and OCI-Ly10 cells in which CARD11 is constitutively activated. In contrast, a high level of ubiquitinated c-Jun was detected in cells with intact CARD11 (OCI-Ly7). Therefore, our study suggests that elevated CARD11 leads to the stabilization and accumulation of c-Jun in DLBCL, which is most likely responsible for aberrant AP-1 activity and induction of AP-1–dependent genes in lymphoma cells.
To study the biological consequences of elevated c-Jun and JunB expression, we treated DLBCL cells with a JNK inhibitor or Jun-specific shRNAs. As we expected, the proliferation of treated cells was reduced, but this effect was much stronger in vivo than in vitro, suggesting that AP-1 induces the expression of genes promoting lymphoma interaction with the microenvironment, which may impact lymphoma growth in vivo. Furthermore, our in vivo study using a xenograft model of DLBCL and our analysis of clinical samples indicated that elevated Jun activity is associated with lymphoma dissemination to extranodal sites, including bone marrow. Indeed, the gene expression profiling of cells with downregulated c-Jun and JunB expression revealed that these transcription factors control multiple genes involved in cell migration, invasion, and metastasis. Among them, we identified matrix metalloproteinase 7, adhesion molecule CD44, vitronectin receptor (ITGAV), fractalkine receptor (CX3CR1), and transcription factor FoxC1, which are all known to promote the invasion and metastasis of solid tumors.25,27,35-40 Intriguingly, the increased expression of ITGAV, FoxC1, and CX3CR1 not only correlated with the elevated c-Jun level but also were enriched in secondary extranodal lymphoma.
There is growing evidence that the crosstalk between neoplastic B cells and components of the microenvironment promote lymphoma growth and progression. Selective expression of chemokines, chemokine receptors, and adhesion molecules can be critical for lymphoma cell homing toward the supporting cells and promote their survival.41 Our study revealed significant changes in the cytokine expression profile upon knockdown of c-Jun and JunB in DLBCL cells. These cells lost their ability to produce IL-6 and IL-10, which are factors that stimulate the growth of neoplastic cells, including non-Hodgkin lymphoma cells.23,42,43 It has been shown that both IL-6 and IL-10 initiate signal transducer and activator of transcription 3 (STAT3) signaling by binding to their surface receptors and activating Janus kinases, which in turn phosphorylate STAT3.44 Consistent with above, Jak/STAT3 signaling is constitutively activated in multiple DLBCL cell lines and patient samples.24,45,46 Although a previous study demonstrated that both cytokines are transcriptional targets of NF-κB,24 we have demonstrated the critical dependence of IL-6 and IL-10 expression on AP-1.
Furthermore, we have identified the requirement of Jun in stroma-induced production of proangiogenic growth factors. Coculture of lymphoma cells with human mesenchymal cells dramatically increased the secretion of VEGF and FGF2. This effect was diminished upon Jun knockdown in lymphoma cells indicating the possible role of Jun transcription factors in angiogenesis. Previous publications suggested that JunB regulates the expression of VEGF in multiple cancer cell lines47-49 as well as in normal fibroblasts.50 However, the latest study reported that selective loss of JunB in stromal cells did not affect tumor angiogenesis,47 suggesting that activation of JunB is essential for malignant cells, but not tumor stroma, to initiate angiogenesis at the metastatic site. This concept would partially explain the higher metastatic potential of lymphoma cells with elevated expression of Jun factors.
Finally, our study revealed that Jun signaling mediates the interaction of lymphoma cells with ECM proteins. Silencing Jun factors in OCI-Ly3 and OCI-Ly10 cells almost completely abolished their adhesion onto fibronectin- and vitronectin-coated plates, suggesting a role of Jun in integrin-mediated adhesion. Consistent with this observation, the gene expression profiling of cells expressing Jun shRNA revealed downregulation of several integrins, including ITGAV. Further analysis with immunoperoxidase tools confirmed that the integrin pathway was a key canonical pathway affected upon Jun silencing in lymphoma cells. Thus, our results strongly point to a critical role of activated Jun signaling in the physical interaction of lymphoma cells with components of the microenvironment.
In summary, we have provided several lines of evidence that CARD11-Jun signaling mediates the interaction of lymphoma cells with the microenvironment and contributes to lymphoma growth and dissemination. Therefore, activated Jun signaling may be one of the major drivers of the aggressive phenotype of DLBCL.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Hans Messner (Ontario Cancer Institute, Canada) for the diffuse large B-cell lymphoma cell lines and Dr Jan Burger (The University of Texas MD Anderson Cancer Center, Houston, TX) for the M2-10B4 bone marrow fibroblasts. The archival tumor specimens were provided by the Anderson Cancer Center Tissue Bank, which is supported by National Institutes of Health, National Cancer Institute grant CA16672.
This work was partially supported by the National Institutes of Health, National Institute of General Medical Sciences grant R01GM065899 and National Institutes of Health, National Institute of Allergy and Infectious Diseases grant R01AI050848 to X.L. and a pilot grant from the Center for Inflammation and Cancer of MD Anderson.
Authorship
Contribution: M.B. conceived and designed the project, performed experiments, analyzed data, and wrote the paper; Y.Z. and K.K. performed experiments; H.H.C. analyzed data and revised the manuscript; M.J.Y. and F.V. provided clinical samples and revised the manuscript; and X.L. analyzed data and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for M.B. is Department of Medicine, The University of Miami, Miami, FL. The current affiliation for Y.Z. is The First Affiliated Hospital, Sun Yat-sen University, Guangzhou, China.
Correspondence: Marzenna Blonska, Department of Medicine, The University of Miami Sylvester Cancer Center, 1400 NW 12th Ave, Miami, FL 33136; e-mail: mblonska@med.miami.edu; and Xin Lin, Department of Molecular and Cellular Oncology, The University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd, Unit 108, Houston, TX 77030; e-mail: xllin@mdanderson.org.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal