To the editor:
Mutations in PRF1, which encodes perforin, were discovered to cause familial hemophagocytic lymphohistiocytosis (FHL) in 1999 and account for 20% to 50% of all FHL cases.1-6 Flow cytometric detection of perforin in peripheral blood natural killer (NK) cells and cytotoxic T lymphocytes was previously studied in a small number of patients by our group and suggested that flow cytometry offered a rapid and sensitive approach for detection of perforin deficiency.7 To more accurately determine the clinical diagnostic accuracy of flow cytometric screening for the detection of PRF1 mutations, we reviewed the reported results of 750 clinical samples submitted to the Diagnostic Immunology Laboratory at Cincinnati Children’s Hospital by the ordering physician for perforin testing between August 2009 and December 2014; genetic sequencing was also performed as directed by the ordering physician. For patients with multiple results, only the first sample with a reportable result was included. Samples tested after bone marrow transplantation were excluded. The flow cytometric measurement of perforin was performed as previously described7 and details are available in the supplemental Data, available on the Blood Web site. Details regarding intra- and interassay variability, establishment of the age-based normal range, genetic sequencing, and statistical analysis are also available in the supplemental Data.
Samples were almost equally distributed between males (n = 383) and females (n = 367). The median age of patients was 5 years (range, 5 days to 66 years). Among the samples, 538 (72%) were observed to have normal perforin expression, 170 (23%) were observed to have low perforin expression, and 42 (7%) had no perforin expression. Of the 212 samples with low or no perforin expression, 111 possessed abnormal sequencing results in PRF1. In total, 166 samples possessed abnormal sequencing results. Forty-eight samples (46 with low or no perforin) were found to have biallelic disease-causing mutations, 26 (18 with low or no perforin) were found to have monoallelic pathological mutations, and 92 (47 with low or no perforin) were found to have one or more variants of uncertain clinical significance, including the A91V variant. Among the 48 patients affected with biallelic disease-causing mutations, 27 patients (56%) had deletion mutations in 1 or 2 alleles and had no detectable protein expression (supplemental Table 3), with 50delT (L17fsX50) being the most common mutation. Biallelic missense mutations were detected in 21 of 48 affected patients and were more likely to yield some protein expression (7 of 21 samples affected by missense mutations).
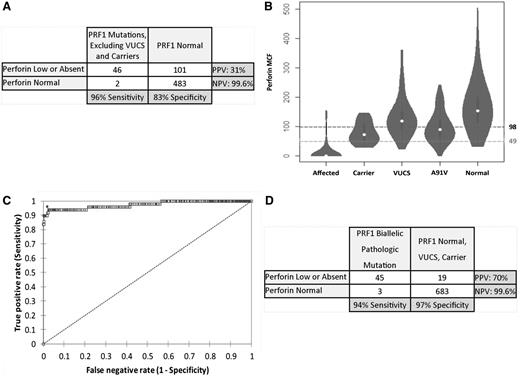
The use of the laboratory-defined normal range to identify patients with biallelic mutations compared with patients with normal sequencing results yielded a sensitivity of 96%, specificity of 83%, positive predictive value (PPV) of 31%, and negative predictive value (NPV) of 99.6% (Figure 1A). However, a considerable proportion of patients with monoallelic PRF1 mutations (69%), variants of uncertain clinical significance (35%), or the A91V variant (60%) and a minority of normal patients (those referred for testing for PRF1 mutations but for whom no mutations were found) (17%) had perforin results below the laboratory-defined lower normal limit of NK cell perforin mean channel fluorescence of 98 (Figure 1B). When we compared patients with biallelic mutations to patients with all other sequencing results, we observed a lower diagnostic accuracy, with sensitivity of 96%, specificity of 76%, PPV of 22%, and NPV of 99.6%. We next performed receiver operating characteristic analysis to define an optimal diagnostic threshold that would identify patients with biallelic pathological mutations with maximum sensitivity and specificity against all other patient groups (Figure 1C). The diagnostic parameters of the test using an optimum NK cell perforin mean channel fluorescence threshold of 49 included a sensitivity of 94%, specificity of 97%, PPV of 70%, and NPV of 99.6% (Figure 1D).
Diagnostic accuracy of flow cytometric perforin screening. (A) Diagnostic accuracy of perforin flow cytometric screening to detect patients with biallelic pathological PRF1 mutations compared with patients with normal sequencing results, using the laboratory-determined normal range for perforin mean channel fluorescence (MCF) in NK cells. (B) Violin plot representation of NK-cell perforin MCF in relation to genetic classification including affected (biallelic disease-causing mutations), carrier (monoallelic mutation with or without additional variants of uncertain clinical significance), variant(s) of unknown clinical significance (VUCS), A91V variant(s), and normal. The laboratory normal control range threshold (perforin MCF, 98) is shown with a dashed line, and the optimum diagnostic threshold obtained by an analysis of receiver operating characteristics (perforin MCF <49) is shown with a dashed line. (C) Receiver operating characteristic curve analysis of perforin MCF showing optimal threshold test characteristic as determined by the Youden index (*). Area under the curve is 0.974. (D) Diagnostic accuracy of perforin flow cytometric screening to detect patients with biallelic pathological PRF1 mutations compared with patients with normal sequencing results, monoallelic mutation with or without additional variants of uncertain clinical significance, and VUCS using the optimal diagnostic threshold established by receiver operating characteristic curve analysis.
Diagnostic accuracy of flow cytometric perforin screening. (A) Diagnostic accuracy of perforin flow cytometric screening to detect patients with biallelic pathological PRF1 mutations compared with patients with normal sequencing results, using the laboratory-determined normal range for perforin mean channel fluorescence (MCF) in NK cells. (B) Violin plot representation of NK-cell perforin MCF in relation to genetic classification including affected (biallelic disease-causing mutations), carrier (monoallelic mutation with or without additional variants of uncertain clinical significance), variant(s) of unknown clinical significance (VUCS), A91V variant(s), and normal. The laboratory normal control range threshold (perforin MCF, 98) is shown with a dashed line, and the optimum diagnostic threshold obtained by an analysis of receiver operating characteristics (perforin MCF <49) is shown with a dashed line. (C) Receiver operating characteristic curve analysis of perforin MCF showing optimal threshold test characteristic as determined by the Youden index (*). Area under the curve is 0.974. (D) Diagnostic accuracy of perforin flow cytometric screening to detect patients with biallelic pathological PRF1 mutations compared with patients with normal sequencing results, monoallelic mutation with or without additional variants of uncertain clinical significance, and VUCS using the optimal diagnostic threshold established by receiver operating characteristic curve analysis.
Our results indicate that clinical flow cytometric screening for perforin deficiency is quite sensitive (greater than 90%) and is associated with a low false-negative rate. Importantly, more than 99% of patients with normal flow cytometric results can be accurately diagnosed as not having perforin deficiency. On the basis of our experience, we recommend that flow cytometric screening be pursued for all patients suspected of having FHL. Testing results are available within a few days and should be helpful in guiding initial treatment decisions and consideration for bone marrow transplantation while awaiting more lengthy confirmatory genetic tests.
The online version of this article contains a data supplement.
Authorship
Contribution: M.A. analyzed data and wrote the manuscript; S.C. contributed to data analysis and reviewed the manuscript; K.Z. supervised the genetic sequencing and interpreted genetic results; A.H.F., J.B., C.G., and J.M.V. developed the assay, supervised sample analyses, and reviewed the manuscript; and R.A.M. designed the analysis, performed statistical analyses, interpreted results, and edited the manuscript.
Conflict-of-interest disclosure: Revenue generated by the clinical testing performed in the Diagnostic Immunology Laboratory funds a proportion of the salaries for A.H.F., C.G., J.V., and J.J.B., and funds the research, development, and validation of clinical tests.
Correspondence: Rebecca A. Marsh, Division of Bone Marrow Transplantation and Immune Deficiency, Cincinnati Children’s Hospital Medical Center, 3333 Burnet Ave, Cincinnati, OH 45229; e-mail: rebecca.marsh@cchmc.org.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal