In this issue of Blood, Guièze et al show that concurrent driver gene mutations are frequent in patients with relapsed/refractory chronic lymphocytic leukemia (CLL) and associated with worse outcome.1 Using deep sequencing, the authors analyzed a panel of known recurrently mutated genes from 3 prospective trials investigating chemo(-immuno-)therapy in relapsed/refractory CLL (see figure). Such targeted sequencing platforms are now in reach of any major medical center.
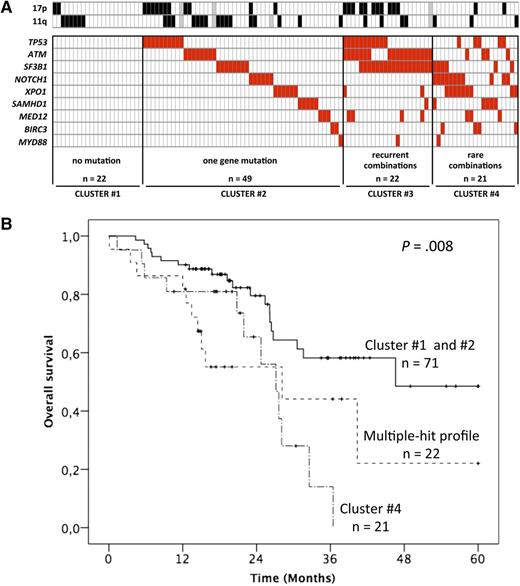
Factors influencing outcome in CLL. (A) The incidence of mutations in relapsed and refractory CLL as summarized by Guièze et al. (B) Kaplan-Meier plots for OS of the mutation groups. See Figures 2 and 5 in the article by Guièze et al that begins on page 2110.
Factors influencing outcome in CLL. (A) The incidence of mutations in relapsed and refractory CLL as summarized by Guièze et al. (B) Kaplan-Meier plots for OS of the mutation groups. See Figures 2 and 5 in the article by Guièze et al that begins on page 2110.
The authors identify patterns of mutations that are in keeping with the previous comprehensive cartography studies.2-5 Because of the advanced stage of relapsed refractory CLL, the incidence of high-risk mutations is high, with >10% for XPO1 and NOTCH1 and >20% for TP53, SF3B1, and ATM. To put these numbers into perspective, a recent study of indolent CLL reported incidences <10% for each of these genes.6
In their careful study, Guièze et al classified the patients based on the absence, single, or multiple occurrence of mutations in this 9-gene panel. It is important to remember that the average number of protein coding gene mutations in CLL is 20,5 and the mutation frequency is associated with IGHV mutation status.
The multiple hit group was associated with a poorer prognosis with regard to progression-free and overall survival (OS). In fact, the patients with >1 mutation in the 9 genes (groups 3 and 4 in Guièze et al) had a significantly poorer outcome, with a median OS of 28.2 and 27.1 months, respectively. With the limited sample size of this study, it is difficult to disentangle the genes’ individual effects from that of the multiple hit combinations.
What are the implications? We tend to simplify the impact of prognostic factors or gene mutations into single dimensions such as presence or absence. It is becoming clear that clonal size and structure,4 convergence,7 and combinations of mutations, as highlighted by this work, will affect outcome. Through advances in sequencing technology, the clonal composition at the genetic level is now relatively simple to investigate, which should not obscure the fact that multiple and much more complex factors including host and additional tumor properties will affect outcome. This immediately raises some of the limitations of the current and comparable studies. The study included controlled trial patients, but treatment was diverse, and the numbers did not allow assessment of the predictive impact of mutations. In general, it is somewhat sobering to acknowledge that, although many prognostic factors have been identified, our arsenal of predictive factors and thus tailored treatments is very limited. In addition, it is more difficult to assess prognostic impact of genetic lesions of patients in relapsed/refractory trials because of sample size and selection bias, which are hard to control for, and also because our definitions of relapsed and refractory disease are relatively crude.
With the emergence of novel treatments targeting the B-cell receptor pathway and the programmed cell death machinery, it will be particularly important to systematically assess the clonal tides produced in the context of selective pressure. Indeed, the analysis of the genetic fingerprint produced by drug selection is highly informative. In current practice, with most patients receiving chemoimmunotherapy, the emergence of ATM, TP53, and SF3B1 mutant cells, as found in the current study, is expected. As we increasingly use drugs with distinctly different mechanism of action, novel mutation targets (eg, phospholipase C-γ and Bruton tyrosine kinase) emerge, which also implies that future platforms will need to cover additional genes.
In summary, the work of Guièze et al casts light on the potential to improve current prognostic models of CLL and should serve as impetus to develop large-scale and meticulous studies of the genetic architecture of CLL, accounting for the dynamic nature of the mutational process.
Conflict-of-interest disclosure: The authors declare no competing financial interests.