Abstract
Background:
MECOM-rearrangements (formerly named EVI1 -rearrangements) are known to be prognostically adverse. Within the WHO classification of 2008, only inv(3)(q21q26.2)/t(3;3)(q21;q26.2)/RPN1-EVI1 were classified as separate entity ("classic"), whereas all other MECOM-rearrangements ("other") were not. In addition, patients (pts) have to be assigned to MDS or AML according to percentage of blasts. As progression from MDS to AML is a continuous process this arbitrary border can be questioned.
Aim:
To evaluate 1) whether MECOM-rearranged pts, regardless of blast count, show a "secondary-type" mutational profile and whether this differs between different types of MECOM-rearrangements. 2) the prognostic impact of the biological and genetic parameters.
Patients and Methods:
We here focus on a cohort of 116 newly diagnosed adult pts with MECOM-rearrangements all investigated by cytogenetics, fluorescence in situ hybridization and cytomorphology. In addition, all pts were analyzed for molecular mutations by next generation sequencing using a 18-gene panel including genes frequently mutated in MDS and AML encompassing also those defined by Lindsley et al. as "secondary-type" mutations (mut) in AML, which are highly specific for secondary AML (Blood 2015). Data was available for: ASXL1 (n=104), BCOR (n=101), CBL (n=112), DNMT3A (n=114), EZH2 (n=111), FLT3 -TKD (n=115), IDH1 (n=116), IKZF1 (n=106), KIT (n=115), NRAS (n=113), PTPN11 (n=111), RUNX1 (n=109), SF3B1 (n=113), SRSF2 (n=113), TP53 (n=116), U2AF1 (n=116), WT1 (n=116), and ZRSR2 (n=113). Variants of unknown significance were excluded from statistical analyses (n=16).
Results:
In our cohort of 116 pts (median age: 66 years, range 21-93) 38 (33%) pts harbored <20% bone marrow blast and therefore were classified as MDS and 78 (67%) ≥20% and therefore were AML. According to WHO classification, 45 (39%) carried "classic" and 71 (61%) "other" MECOM-rearrangements. Most common was inv(3)(q21q26) (n=35; 30%), followed by t(3;21)(q26;q22) (13; 11%), t(3;8)(q26;q24) (12; 10%) and t(2;3)(p14~p21;q26) (11; 10%), t(3;3)(q21;q26) (10; 9%), inv(3)(p24q26) (9; 8%), and t(3;21)(q26;q11) (7; 6%). All others were present in less than 5 cases. We found mut in SF3B1 (27%), NRAS (22%), ASXL1 (18%). PTPN11 and SRSF2 (17% each), RUNX1 (15%), DNMT3A (11%), and TP53 (10%). Mutations in the remaining genes were present in less than 10%. In total, 60% of the pts showed at least one of the "secondary-type" mut. Only nine pts (8%) carried no mut.
MDS pts harbored more frequently SF3B1 mut than AML pts (45% vs 19%, p=0.007). Overall, "secondary-type" mut were much more frequent in MDS than in AML (82% vs 49%; p=0.001). No differences with respect to clinical parameters or survival were seen.
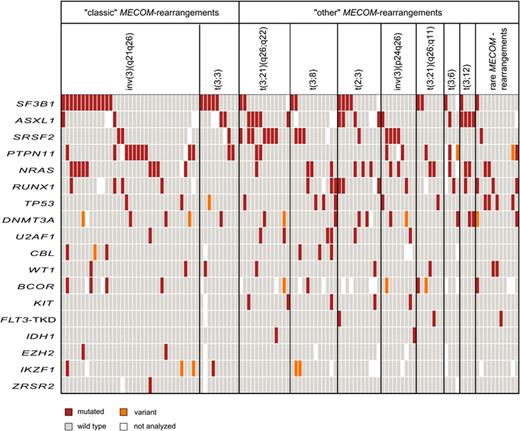
Separating the cohort into "classic" and "other" rearrangements we found "classic" harboring more often SF3B1 mut (40% vs 19%, p=0.019) and PTPN11 mut (28 vs 11%; p=0.037) and less frequent SRSF2 mut (7% vs 23%, p=0.037) compared to "other" pts. No other differences were seen. However, diverse mutation patterns were detected between the distinct rearrangements (Figure 1). Pts with t(3;8) carried 60% CBL and 25% TP53 mut, whereas t(3;3), t(2;3), and t(3;12) yielded none of these mut. Furthermore, t(3;21)(q26;q22) harbored 54% SRSF2 and 39% ASXL1 mut, whereas inv(3)(q21q26) carried only 6 and 7%, respectively, and t(3;21)(q26;q11) carried none of these. Moreover, PTPN11 mut was present in 30% of inv(3)(q21q26) but not in t(3;8), t(2;3), t(3;6), and t(3;12). Further, IDH1 mut were carried by 50% of t(3;21)(q26;q22) and inv(3)(p24q26), but in none of the other pts.
An extremely poor prognosis was observed in patients with NRAS mut and TP53 mut compared to pts without these mut (8 vs 23 months; p=0.002, 6 vs 16 months, p<0.001, respectively). Of note, the cumulative number of mutations had a negative impact on survival: pts with 0-1 vs. ≥2 mut, median OS 41 versus 9 months, p=0.001.
Conclusions
1. A threshold of 20% bone marrow blasts does not separate patients with MECOM-rearrangements according to clinical or genetic parameters or outcome.
2. "Classic" MECOM-rearrangements do not differ from "other" ones according to clinical parameters or outcome.
3. However, "classic" and "other" MECOM-rearrangements show different profiles of additional mutations.
4. Furthermore, additional molecular markers lead to better separation of MECOM-rearranged patients with regard to outcome.
Alpermann:MLL Munich Leukemia Laboratory: Employment. Haferlach:MLL Munich Leukemia Laboratory: Employment, Equity Ownership. Fasan:MLL Munich Leukemia Laboratory: Employment. Schindela:MLL Munich Leukemia Laboratory: Employment. Kern:MLL Munich Leukemia Laboratory: Employment, Equity Ownership. Haferlach:MLL Munich Leukemia Laboratory: Employment, Equity Ownership.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal