Abstract
Background: Whole genome sequencing (WGS) identified activating CXCR4WHIM somatic mutations in nearly 30% of patients with Waldenstrom's Macroglobulinemia (WM) (Blood 123(11):1637-46). Both nonsense and frameshift CXCR4WHIM mutations occur in WM, with over 30 different types of mutations described within the regulatory carboxyl-terminal domain of CXCR4. CXCR4WHIM mutations almost always occur with activating MYD88 mutations, and impact both disease presentation and treatment outcome (Blood 123(18):2791-6; NEJM 372(15):1430-40.). The clonal architecture of CXCR4WHIM mutations relative to MYD88 mutations and their role in disease evolution remains to be clarified.
Methods: We used Sanger sequencing and highly sensitive AS-PCR assays that we developed for the most common CXCR4WHIM mutations (S338X C>A and C>G) to evaluate for CXCR4WHIM mutations. In conjunction with an AS-PCR MYD88L265P assay that we previously developed (Leukemia 28(8):1698-707), we also profiled tumor samples for MYD88L265P and CXCR4S338X mutations in 164 WM, 12 IgM MGUS, 20 MZL, 32 CLL, 14 MM, 7 non-IGM MGUS patients, and 32 healthy donors. Next generation transcriptome sequencing data was also performed for validation in select cases.
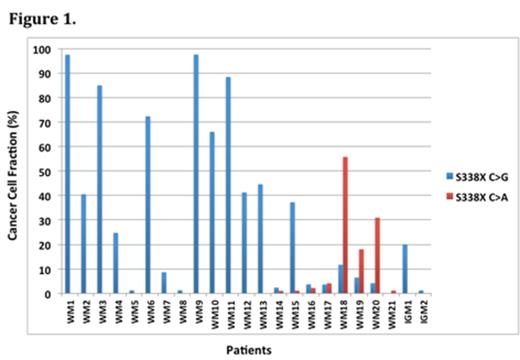
Results: AS-PCR detected CXCR4S338X mutations in WM and IgM MGUS patients not revealed by Sanger sequencing. By combined AS-PCR and Sanger sequencing, CXCR4WHIM mutations were identified in 44/102 (43%), 21/62 (34%), 2/12 (17%), and 1/20 (5%) untreated WM, previously treated WM, IgM MGUS and MZL patients, respectively, but not in CLL, MM, non-IGM MGUS patients or healthy donors. Cancer cell fraction analysis in WM and IgM MGUS patients showed CXCR4S338X mutations were primarily subclonal, with highly variable clonal distribution (median 35.1%, range 1.2%-97.5%; Figure 1). Sanger sequencing identified 3 patients with multiple CXCR4 mutations, which were shown to be compound heterozygous by TA cloning and sequencing of at least 40 clones. The addition of AS-PCR to the Sanger sequencing results also revealed multiple CXCR4WHIM mutations in many individual patients that included homozygous and compound heterozygous mutations that were validated by next generation sequencing that offered a median of 5,819 (range 5,217-12,235) reads that overlapped the mutated loci.
Conclusions: Taken together, these findings show that CXCR4WHIM mutations are more common in WM patients than previously revealed by WGS or Sanger sequencing. Moreover, CXCR4 mutations are primarily subclonal supporting their acquisition after MYD88L265P in WM oncogenesis. The exclusive finding of frameshift and nonsense but not missense variants within the carboxyl-terminal domain of CXCR4 suggests that significant selection pressures exist for activating mutations within the WM clone. Lastly, multiple CXCR4WHIM mutations are common in WM patients indicative of targeted genomic instability within the carboxyl-terminal domain of CXCR4.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal