Abstract
Introduction
Exome sequencing can detect somatic mutations at an unprecedented scale. However, high false-positive rates due to multiple technical contributors affecting signal-to-noise ratio is an unsolved inherent problem. Furthermore, the current literature does not consistently assess or report the allelic burden of detected and scrutinized mutations, potentially leading to focus on irrelevant mutations. We hypothesized that this issue can be addressed by a structured usage of allele frequencies in combination with detected mutations. Employing improved techniques to assess the molecular signature shortly after diagnosis could also result in improved risk stratification. Here, we present proof-of-principle that identification of relevant mutations, and extent of clonality, can be accomplished by pairing hypothetical allelic burden deduced from sequencing reads to leukemic burden in individual samples.
Methods
7 samples were used for whole exome sequencing derived from bone marrow aspirated from two different patients. Of these, 4 were leukemia samples (diagnostic and two relapse samples of one T-cell/myeloid mixed-phenotype leukemia and a diagnostic from AML, M0), reflecting different clinical situations, and 3 samples served as controls (keratinocytes, and fibroblasts in the first, remission in the second) to include different clinical situations. Raw data processing, mutations analysis and downstream analysis of sequencing data was performed as described earlier (Hansen & al., MethodsX 2015, Hansen & al., Br J Haematol. 2015), following GATK Best Practice workflow and MuTect default parameters. Cluster analysis was utilized to mathematically divide somatic SNVs into read frequency clusters on the basis of squared Euclidean distance, thus enabling the retrieval of mean allelic burden and expected allele frequencies juxtaposed with flow cytometry-derived leukemic blast percent. The initial condition of the classification model was default set to accommodate separation of background (cluster 1: noise or low read), heterozygous mutations (cluster 2), and an outlier bin for possible homozygous mutations (cluster 3).
Results and discussion
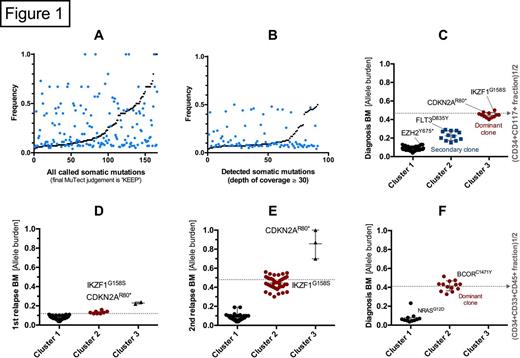
In order to reference the leukemic burden of a given sample we employed data from immunophenotyping as a surrogate marker for malignancy. Without filtering of detected mutations by exome sequencing the partly stochastic nature of the signal is evident (fig. 1A). An exponential-like continuum of non-rejected mutations is found when these are sorted according to frequencies (black points), making it difficult to deduce any clonal nature of the malignancy, and to evaluate the validity of possibly relevant mutations found. Using a minimum depth of coverage threshold of 30 the signal-to-noise ratio is increased (fig. 1B), and, subsequently, two clones were resolved at diagnosis (fig. 1C) by cluster analysis. This resolution decreases at lower tumor burden showing expected sensitivity towards lower allelic reads (fig. 1D), although driver mutations of the persistent clone could be detected. At second relapse only one distinct sAML phenotyoe clone and, now homozygous, CDKN2 AR80* could be detected (fig. 1E) with the mutations of secondary clone present at diagnosis (cluster 2) lost due to selective pressure from treatment, i.e. FLT3D835Y. The observations are backed by another diagnostic sample from a patient with AML with NRAS and BCOR mutations (fig. 1F), also with a distinct clone. Here, too much emphasis could easily be given to NRASG12D if the frequency had not been assessed.
Conclusion
We have addressed the pertinent question regarding false positive observations arising from deep sequencing or emphasizing mutations found in the low allele frequency fractions. From this dataset we have, despite the low number of samples, accomplished to suggest a formalized approach for single sample mutational analysis. As a consequence, we can now show that malignant clones with high tumor burden can be resolved semi-spatially by sequencing, generally applicable to a wide range of clinical settings. We conclude that this approach is amenable in single patient situations. While further studies are needed to ultimately test the applicability of this approach in the clinical settings, the perspectives to this observation become evident as sequencing depth and cost continue to develop in an inversely correlated manner.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal