Abstract
Introduction:
Favorable risk acute myeloid leukemia (AML) is defined by recurrent genetic abnormalities including core binding factor rearrangements such as inv(16) and t(8,21), normal cytogenetics with isolated mutations of NPM1, or bi-allelic mutations of CEBPA. Approximately 60% of patients are cured with standard 7+3 induction and cytarabine consolidation, which is comparable to survival rates of patients who receive allogeneic stem cell transplant (SCT). However, 20-30% of patients treated with chemotherapy still relapse, and relapsed disease remains the leading cause of death in this group. Even if relapsed patients achieve CR2 with salvage chemotherapy, their survival with allogeneic SCT is reduced compared to patients transplanted in CR1. Thus, predicting which favorable risk AML patients are more likely to relapse after chemotherapy would help guide therapy and improve patient outcomes. Recent publications have proposed that additional mutations in genes such as IDH1, IDH2, or DMT3A may impact relapse risk, but reports are conflicting. In addition, studies using minimal residual disease to evaluate disease burden after induction and consolidation has also been shown to predict relapse. We have used high throughput next generation sequencing (NGS) as diagnostic panel for AML at our institution for the past 2 years. This panel looks for mutations in 42 different genes known to be associated with acute leukemias and can quantitatively evaluate genetic MRD at a sensitivity of about 0.1%. We are analyzing favorable risk AML patients in an effort to identify additional mutations that predict relapse and in the process of evaluating the ability of this panel to evaluate MRD.
Methods:
Clinical samples were obtained with informed consent and with the approval of our institutional IRB. A targeted NGS panel was designed using multiplexed Ion AmpliSeq Designer (Life Technologies) software to amplify and sequence 42 genes relevant to hematopoietic malignancies. 20ng of DNA from bone marrow or blood was used to generate amplicon-based libraries that were sequenced using an Ion Torrent PGM. Bioinformatics analysis was performed using the Torrent Suite v.3.2 pipeline. Open source programs and lab-developed algorithms were used for variant annotation and mutation prediction.Patients with favorable risk AML diagnosed within the past two years had sequencing data available in their medical records. For patients diagnosed >2 years ago, archival samples were obtained and DNA extracted from isolated mononuclear cells or formalin-fixed paraffin-embedded tissue according to standard protocols.
Results:
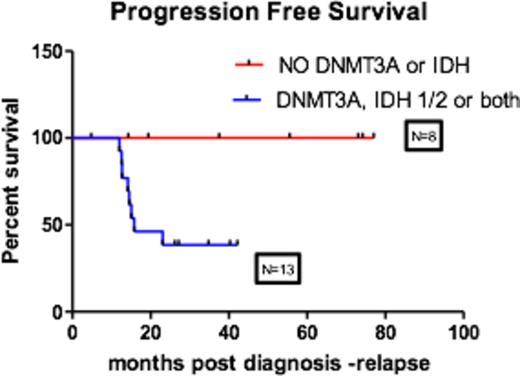
We identified 57 patients with favorable risk AML diagnosed at OHSU over the past five years. 48 had enough biopsy material for genetic analysis. Of the 48, 17% had t (8;21), 29% had inv(16), 46% were NPM1+, and 8% had CEBPA mutations. Seven patients received transplant in CR1 for either residual disease or physician discretion. Of the remaining 41 patients, 11 patients relapsed (26%) and 8 of the 11 were NPM1+. Of the 8 relapsed NPM1+ patients, all had additional DNMT3A R882 mutations, IDH1/2 mutations, or both. 13 NPM1+ non-relapsed patients have been evaluated by sequencing to date and only 5 of 13 in the non-relapsed group had additional mutations in DNMT3A and/or IDH1/2(P=0.0185), however only 2 of the 5 had the R882 DNMT3A mutation. No DNMT3A, IDH1 or IDH2 mutations were identified in patients with t(8;21), inv (16) or biallelic CEBPA AML.
Conclusions:
Although our numbers are small, the presence of the R882 DNMT3A mutation appears to increase the risk of relapse in NPM1+ patients (Figure 1). Other DNMT3A point mutations do not seem to impact the risk of relapse and should be considered separately for relapse risk. Complete analysis and evaluation of MRD is underway.
Dunlap:Oregon Health & Sciences University: Employment. Druker:Bristol-Myers Squibb: Research Funding; McGraw Hill: Patents & Royalties; Oncotide Pharmaceuticals: Research Funding; Millipore: Patents & Royalties; Leukemia & Lymphoma Society: Membership on an entity's Board of Directors or advisory committees, Research Funding; Aptose Therapeutics Inc.: Consultancy, Equity Ownership, Membership on an entity's Board of Directors or advisory committees; ARIAD: Research Funding; AstraZeneca: Consultancy; Blueprint Medicines: Consultancy, Equity Ownership, Membership on an entity's Board of Directors or advisory committees; CTI Biosciences, Inc.: Consultancy, Equity Ownership, Membership on an entity's Board of Directors or advisory committees; Cylene Pharmaceuticals: Consultancy, Equity Ownership, Membership on an entity's Board of Directors or advisory committees; Fred Hutchinson Cancer Research Center: Research Funding; Henry Stewart Talks: Patents & Royalties; Gilead Sciences: Consultancy, Membership on an entity's Board of Directors or advisory committees; Novartis Pharamceuticals: Research Funding; Molecular MD: Consultancy, Equity Ownership, Membership on an entity's Board of Directors or advisory committees; Oregon Health and Science University: Patents & Royalties; Roche TCRC, Inc.: Consultancy, Membership on an entity's Board of Directors or advisory committees; Sage Bionetworks: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal