Abstract
Background. Recently, the first genome-wide association study (GWAS) for multiple myeloma (MM) survival was performed. It identified 16p13 as a candidate region for germline risk variants influencing survival1. The Utah study contributed to a successful replication of this region. Here, we perform a GWAS of MM survival in 318 MM cases residing or being treated in Utah to identify additional SNPs associated with MM prognosis.
Methods. Utah cases were treated in the Huntsman Cancer Hospital (HCH) and/or Intermountain Healthcare systems, and were ascertained either though the HCH or the Utah Cancer Registry. Cases were genotyped on the Illumina 610Q high-density SNP array containing over 600,000 variants across the genome, imputed to the 1000 Genomes sequence data. Only imputed genotypes with information content >0.7 and certainty >0.9 were considered. For all variants (genotyped or imputed) with minor allele frequency of ≥2%, we estimated genome-wide hazard ratios using Cox models to compare survival between MM carriers of major or minor SNP alleles. Models were adjusted for sex, age at diagnosis, and principal components to adjust for possible population stratification as covariates. Disease stage and treatment were considered in sensitivity analyses to ensure that the hazard ratios were robust to these important factors. All Cox analyses were performed in R and Kaplan-Meier estimates of the survival function were graphed. Suggestive regions (p <1×10-6) were taken forward to replication in the UCSF study (352 patients). Combined analyses using METAL were calculated, including Cochran's Q test of heterogeneity.
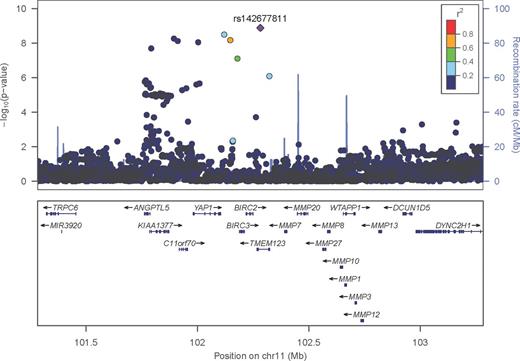
Results. Of 16 regions with p <1×10-6, we identified four regions on chromosomes 9, 10, 11 and 16 that were replicated in the UCSF data (p =0.007 to 0.06): (1) 9q22 rs12346172, p =5.7×10-7, HR=7.7; (2) 10p12.2 rs67205241, p =8.0×10-7, HR=3.1; (3) 11q22 rs142677811, p =1.3×10-9, HR=8.6 (see Figure 1); and, (4) 16q24 rs87814765, p =8.5×10-7, HR=2.1. These regions maintain genome-wide significance (rs142677811, p =2.1×10-8) or near-significance in a Utah-UCSF Meta analysis. Estimates were not sensitive to differences in staging or treatment.
Conclusions. We observed that germline variants near genes PTCH1 (9q22.3; tumor suppressor in the patched gene family), EBLN1 (10p12.31), BIRC2 and BIRC3 (inhibitors of apoptosis; 11q22), YAP1 (oncogene in the Hippo signaling pathway; 11q13) and SLC7A5 (16 q24.3; an L-type amino-acid transporter recently associated with poor prognosis in MM2) were associated with decreased survival among MM patients. Somatic mutation and copy number loss for BIRC2/3, proteins in the inhibitor of apoptosis family, are frequently detected in lymphoid malignancies3. Nuclear ABL1 triggers cell death through its interaction with the Hippo pathway coactivator YAP1 in normal cells; it has been shown that low YAP1 levels prevent nuclear ABL1-induced apoptosis in hematological malignancies4.
References:
1. Ziv E, Dean E, Hu D, et al. Genome-wide association study identifies variants at 16p13 associated with survival in multiple myeloma patients. Nat Commun. 2015 Jul 22;6:7539.
2. Isoda A, Kaira K, Iwashina M, et al. Expression of L-type amino acid transporter 1 (LAT1) as a prognostic and therapeutic indicator in multiple myeloma. Cancer Sci. 2014 Nov;105(11):1496-502.
3. Yamato A, Soda M, Ueno T, et al. Oncogenic activity of BIRC2 and BIRC3 mutants independent of nuclear factor-κB-activating potential. Cancer Sci. 2015 Jun 19.
4. Cottini F, Hideshima T, Xu C, et al. Rescue of Hippo coactivator YAP1 triggers DNA damage-induced apoptosis in hematological cancers. Nat Med. 2014 Jun;20(6):599-606.
Plot of negative log p significance values for association statistics at the chromosome 11q22 region, bp 101282470-103282470 (h.g. build 19) in the Utah study, using locuszoom software (http://csg.sph.umich.edu/locuszoom/). The top associated SNP (rs142677811) is colored in purple and the remaining SNPs are colored according to linkage disequilibrium values (r2) with the top SNP.
Plot of negative log p significance values for association statistics at the chromosome 11q22 region, bp 101282470-103282470 (h.g. build 19) in the Utah study, using locuszoom software (http://csg.sph.umich.edu/locuszoom/). The top associated SNP (rs142677811) is colored in purple and the remaining SNPs are colored according to linkage disequilibrium values (r2) with the top SNP.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal