Abstract
Background:
Primary testicular diffuse large B cell lymphoma (DLBCL) is an uncommon malignancy portending a poor prognosis with increased risk of central nervous system disease. Phenotypically, most primary testicular lymphomas have a non-germinal center B-cell like (non-GCB) origin. To identify the genetic characteristics of testicular DLBCL, we evaluated DNA copy number and mutational profiling using SNP array and a next generation targeted sequencing platform.
Methods:
Twelve cases of testicular DLBCL with patient consent for tissue specimens and sufficient tumor tissue were retrospectively identified. Cell of origin was determined by Hans immunohistochemistry (IHC) model. We performed a custom, targeted deep-sequencing assay of 585 cancer genes (HemePACT) on matched tumor and normal pairs. Barcoded pools were sequenced on Illumina HiSeq 2500 to 500-1000x coverage per sample Sequencing was compared to a matched normal tissue control (N=10) if available or alternatively a pooled normal tissue control. We excluded all mutations either present at a high variant allele frequency in the matching germline samples, present in two databases of inherited variants (DBSNP and 1000 genomes) or present in one databases of inherited variants and absent from COSMIC. We evaluated copy number and allelic imbalance with an Affymetrix OncoScan SNP-array. IHC was performed for select genes. Results were compared to a panel of non-testicular DLBCL previously described (N=78).
Results:
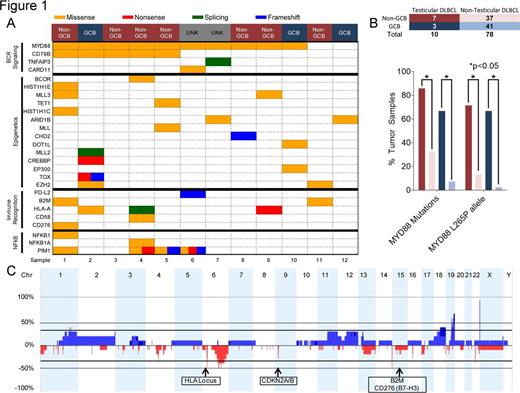
The median age of the patients was 55.1 years (range 21.9-77.9). Patients had clinical stage IE (50%) and IV (50%) disease. All samples were sequenced from pre-treatment biopsies. Eleven of 12 patients were initially treated with R-CHOP chemotherapy, intrathecal methotrexate and radiation. Treatment history for one patient was unknown. We identified 124 mutations in 12 cases of testicular DLBCL. The most common mutation was MYD88 occurring in 10/12 patients (83%) with 6 mutations in non-GCB and 2 mutations in GCB (Fig 1A). The MYD88 L265P allele was most frequent and occurred in 9/12 patients (75%). The median MYD88 L265P variant allele frequency was 0.36 (range 0.07-0.51) with normal copy number status at that loci. In contrast, MYD88 mutations were less frequent in DLBCL without testicular involvement, 12/37 (32%) non-GCB and 3/41 (7%) GCB DLBCL, p<0.05 by Fisher's t-test (Fig 1B). Furthermore, mutations in CD79B were significantly more common in testicular DLBCL (5/12, 42%) versus non-testicular DLBCL (7/78, 9%). Concurrent mutations affecting the BCR receptor pathway was noted in 10/12 patients (83%): CD79B (5/12, 42%), TNFAIP3 (1/12, 8%), CARD11 (1/12, 8%) (Fig 1A). Frequency of TNFAIP3, CARD11 was not statistically significant between testicular versus non-testicular DLBCL. Other commonly mutated pathways include epigenetics (10/12, 83%) and immune recognition (6/12, 50%). Deregulation of immune recognition was noted by HLA-A (3/12, 25%) and beta-2-microglobulin (2/12, 17%) mutations as well as loss of HLA locus in 8/10 samples (80%) (Fig 1C). IHC revealed 6/10 (60%) cases with no MHC Class I expression of the tumor cells. MHC Class I negative cases also lacked B2M expression (3/6) or displayed mislocalization of B2M to the cytosol (3/6). PD-L1 was expressed by lymphoma cells as assayed by IHC in 1/10 (10%) cases but no amplification or mutation was identified. No copy gains of the 9p24.1, PD-1, PD-L1, PD-L2 locus were identified via HemePACT or SNP array. Although mutations in CDKN2A/B were not identified in HemePACT, SNP array confirmed loss of CDKN2A/B at the 9p21 loci in 3/10 cases (30%).
Conclusion:
Targeted genomic sequencing and SNP array analysis have identified a distinctive genetic pattern with alterations of MYD88, BCR pathway mutations, and immune recognition deficiency in testicular DLBCL compared to non-testicular DLBCL. These findings may have implications in guiding the design of future treatment strategies for testicular DLBCL.
Younes:Novartis: Research Funding; Janssen: Honoraria; Johnson and Johnson: Research Funding; Takeda Millenium: Honoraria; Seattle Genetics: Honoraria, Research Funding; Sanofi-Aventis: Honoraria; Bayer: Honoraria; Bristol Meyer Squibb: Honoraria; Curis: Research Funding; Celgene: Honoraria; Incyte: Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal