Abstract
Background: TCLs are an uncommon, heterogeneous group of neoplasms with no consensus on optimal treatment and human 5-year survival rates <20-30%. The canine provides a potentially attractive model to study TCL in part given their spontaneously occurring cancers, intact immune system, and phylogenetic resemblance to humans. Furthermore, approximately 1/3 of all lymphomas in canine are TCL (cTCL). Previous research from our group (ASH 2014, #74755) identified 118 differentially expressed genes by RNA seq analyses comparing canine PTCL with normal canine lymph node, and PI3K, GATA3, GRB2 and PPARG as candidate biomarker genes by canonical pathway and network analyses. We aimed to further interrogate the canine as a model by histologic review and detailed genomic examination of cTCL.
Methods: We evaluated de novo cTCL with immunohistochemistry (IHC) and next generation DNA sequencing for a priori genes. IHC on canine TCL was evaluated with human antibodies against CD5, CD79a, Ki67, CD3, CD4, CD8 and CD30. Canine DNA was extracted from 14 fresh frozen tissue samples and 2 paraffin embedded blocks using the QIAamp DNA Mini Kit. Utilizing the human Cancer Hotspot Panel v2 (hCHPv2), a custom expanded panel of 68 genes actively expressed in lymphoma tumor cells was created to screen cTCL for mutations. COSMIC database and PubMed was used to identify common variants expected to be present. Targets from the hCHPv2 were converted from the human genome (hg19) to positions in the canine genome (canFam3) using liftOver (UCSC Genome Browser). Following targeted amplification using the custom canine library, DNA sequencing was performed with the Ion Personal Genome Machine resulting in 4,527,638 total reads with an average length of 229 bases and 708x coverage per sample.
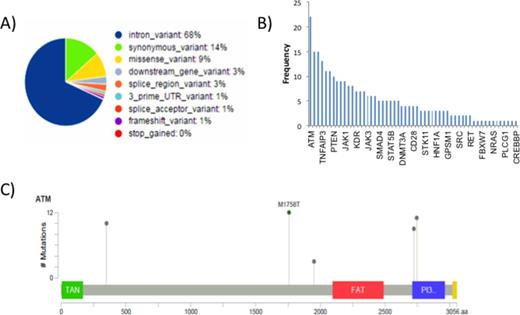
Results: For IHC, we examined 10 primary cTCL cases utilizing human antibodies. The cTCL cases staining patterns included: 100% were CD79a negative; 80% were CD5+; Ki67 was variable; while the remaining multiple markers did not react to human antibodies. We subsequently evaluated 16 primary cTCL tumor tissue samples using DNA sequencing. There were 331 unique variants and 1474 total variants; each sample had an average of 92 variants. The most prevalent coding consequences mutations were intron variations (68%), followed by synonymous (14%) and missense variations (9%) (Fig. 1A). The most prevalent mutations were found in ATM, KIT, ERBB4, TNFAIP3, and TET2 (Fig. 1B). ATM has been implicated as a tumor suppressor and mutations have been described on a case basis in human thymic and mantle cell lymphomas, however, not in TCL. Furthermore, the majority of ATM variants identified in our analysis were non-coding or synonymous. In ATM, there were 22 mutations with 17 found as introns. The 5 coding mutations had only one of which was missense (M1758T), the others were silent (Fig. 1C). Conversely, the most prevalent variants found in the coding region were found in SMO, TP53, TNFAIP3, and TET2. Of all coding variants, 15 missense variants in TNFAIP3, JAK2, MYC, MET, SMO, DNMT3A, RB1, PIK3CA, TP53, and ERBB2 appeared to be deleterious through bioinformatics analysis. Additionally, a frameshift variant in CDH1 resulted in a 3 bp deletion that has not been described in dbSNP. Finally, mutations in KIT, TNFAIP3 and TET2 have been described in canine and human TCL with varying frequency.
Conclusion: To the best of our knowledge, this represents one of the first genomic comparative oncology analyses conducted in TCL. Collectively, the DNA sequencing analysis in cTCL identified genomic similarities and novel mutations that may help unearth new oncogenic pathways in human TCL (e.g., ATM). Furthermore, the deleterious frameshift and missense mutations identified in this study as from TNFAIP3 and CDH1 are novel, of which has not been described in prior research. Continued investigation is needed towards the enhanced delineation of protein expression of cTCL and examination of the functional impact of genomic perturbations identified in cTCL in comparison to human TCL.
Next generation DNA sequencing of canine TCL . A. Coding consequences by variant type. B. All gene variations by frequency C. Mutation map of ATM.
Next generation DNA sequencing of canine TCL . A. Coding consequences by variant type. B. All gene variations by frequency C. Mutation map of ATM.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal