Abstract
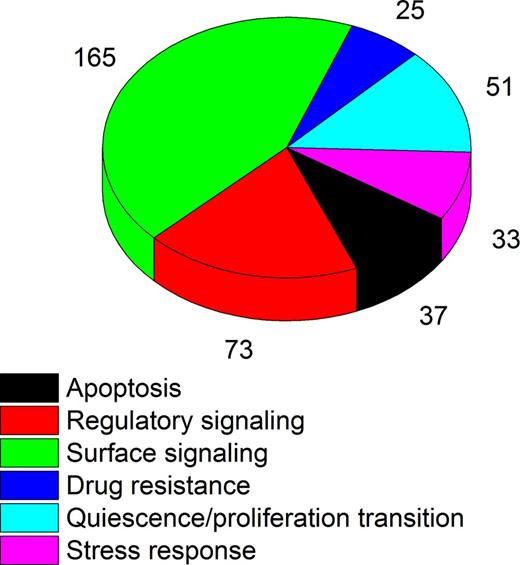
Post-translational modifications of proteins are increasingly recognized as key regulators of functional complexes assembly and cellular enzymes activity, processes that control both normal and aberrant cell growth and development. To enable comprehensive, sensitive and quantitative analysis of these processes, we adapted recently developed high-efficiency nanoscale multidimensional liquid chromatography with high-resolution Orbitrap mass spectrometry. Termed accumulated ion monitoring (AIM), this mass spectrometry method achieved nearly 7 orders of magnitude of quantitative accuracy and absolute limit of detection of 600 molecules per scan, enabling the study of rare cell populations (Fig. 1). We leveraged AIM mass spectrometry to develop a panel of 1583 synthetic reference peptides, based on global and published proteomics maps of normal human and leukemia cells. This Quantitative Cell Proteomics Atlas (QCPA) profiles 384 key effectors of cell surface signaling, proliferation, quiescence, stress response, and epigenetic control of gene expression (Fig. 2). QCPA enables unprecedented accuracy and sensitivity for the functional analysis of rare cells, with focus on protein regulation via phosphorylation, acetylation and methylation, as well as on concentration (http://alexkentsis.net/qcpa). By using functional proteomics profiling of primary human CD34+ and acute myeloid leukemia (AML) cells, we identify new pathways controlling erythrocyte differentiation and AML therapy resistance, as facilitated by a newly developed program ProteoModlR (http://github.com/kentsisresearchgroup/ProteoModlR). AIM mass spectrometry and QCPA functional proteomics are rapidly adaptable and generalizable tools for the investigation of regulatory and epigenetic signaling in normal and diseased cells.
Limits of detection and quantitation for a serially diluted synthetic peptide from human Myocyte-specific Enhancer Factor 2C.
Limits of detection and quantitation for a serially diluted synthetic peptide from human Myocyte-specific Enhancer Factor 2C.
Bauer:Editas Medicine: Consultancy; Biogen: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal