Abstract
Background:
The DNA methyltransferase inhibitors, 5-azactidine and decitabine, are presumed to reverse aberrant methylation in tumor cells from patients with myelodysplastic syndromes (MDS), often resulting in hematologic improvement and even temporary eradication of the malignant clone. This suggests a causative role for abnormal DNA methylation in the development and progression of MDS.
We hypothesize that patterns of DNA methylation associated with somatic mutations may serve as clinical biomarkers. We seek to determine how such somatic mutations in MDS alter the pattern of 5 methylcytosine (5mC) at key regulatory sites in the human genome. Since 5mC constitutes only 1% of total DNA bases, enrichment methods have been developed that are more cost-effective and time efficient compared to whole genome sequencing. Here we compared a novel targeted DNA enrichment approach, bisulfite padlock probes (BSPP), to a more standard enrichment method, reduced representation bisulfite sequencing (RRBS), to determine which method offers broader and more relevant coverage of the methylome in MDS tumor cells and K562 DNA-edited cell lines.
Methods:
We selected eight patient samples harboring somatic mutations that we anticipated would yield aberrant methylation, including mutations in TET2, IDH1, IDH2 and DNMT3A. We performed BSPP and RRBS on genomic DNA purified from bone marrow mononuclear cells. We then utilized the CRISPR/Cas9 approach to introduce mutations into the coding region in K562 myeloid lineage cells, creating DNA-edited cell lines with knockout mutations. We performed BSPP and RRBS on wild-type and mutant TET2 and DNMT3A myeloid cell lines followed by massively parallel sequencing. The resulting sequencing files were quality and adapter trimmed before being mapped to the bisulfite genome using BisReadMapper (Diep et. al 2012). Exploratory analyses including methylation beta value distribution calculation was carried out using the ÒRnBeadsÓ package for R version 3.2.1.
Results:
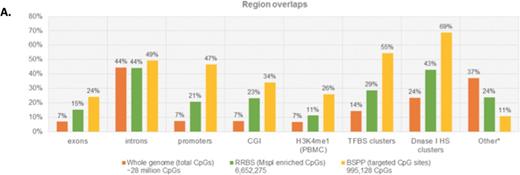
Among the eight patient samples, BSPP covered a quarter as many CpG sites as RRBS when examining regions sequenced in all samples. However, the sites captured by BSPP were sequenced with greater depth and had a higher alignment rate compared to RRBS. Only 3% of RRBS CpG sites were covered by BSPP, representing 12% of BSPP CpG sites. Concordance for 5mC in the regions covered by both techniques was high (R2 = 0.876). Compared to RRBS, genomic coverage beyond CpG islands was much greater with BSPP capture (Figure 1). Enrichment for CpG sites captured by BSPP was particularly seen in promoters, DNAse hypersensitivity sites and transcription factor binding sites.
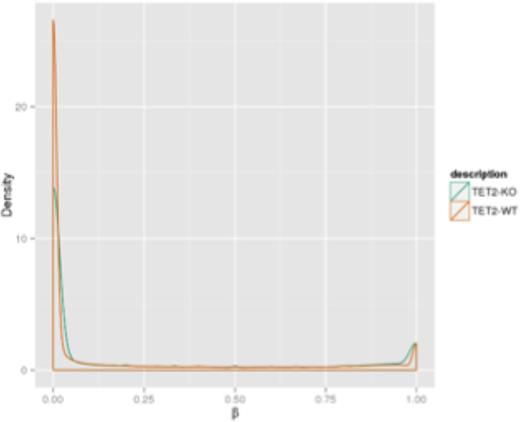
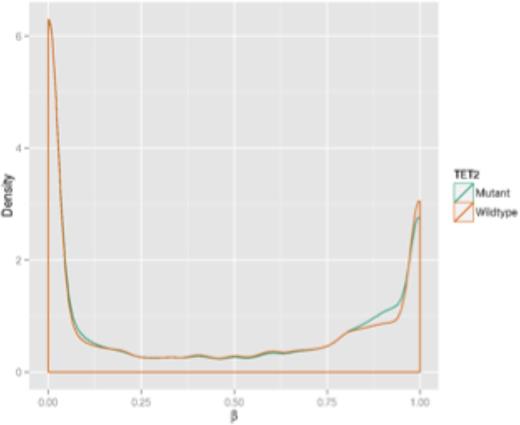
Mutant versus non-mutant methylation beta value density graphs showed similar methylation distributions when comparing data obtained from the patient samples and the K562 cell lines (Figure 2). Methylation values from BSPP data showed greater density distribution in hypomethylated regions among the patient samples and cell lines. Methylation values from RRBS data showed greater density distributions in hypermethylated regions.
Discussion:
RRBS approaches have failed to find pretreatment patterns of methylation associated with response, possibly due to inadequate coverage of 5mC distribution. We were able to obtain enhanced genomic coverage of the methylome utilizing BSPP compared to RRBS on both MDS patient samples and myeloid cell lines. One of the drawbacks of RRBS is its failure to capture areas with lower CpG densities that are potentially relevant to MDS pathobiology. Unlike RRBS, the padlock probes are not limited to regions adjacent to recognition sites of the restriction enzymes, allowing for parallel capture of a much broader range of genomic targets. We assessed methylation value distributions obtained by both approaches, and indeed, BSPP showed greater density distribution in hypomethylated regions in contrast to RRBS. Future work will focus on determining which of these changes in 5mC will prove the greatest predictor of response in MDS patients.
Conclusions:
We have shown that BSPP captures broader coverage of different regulatory regions of the genome compared to a more standard enrichment bisulfite sequencing techniques, RRBS. We have shown that BSPP is able to capture the variance in 5mC between TET2 and DNMT3A mutant and wild-type MDS patients, as well as in K562 DNA-edited cell lines.
Bejar:Genoptix Medical Laboratory: Consultancy, Honoraria, Patents & Royalties: MDS prognostic gene signature; Celgene: Consultancy, Honoraria; Alexion: Other: ad hoc advisory board.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal