Abstract
Background: Multiple myeloma (MM) is a lymphoproliferative disease characterized by the clonal expansion of neoplastic plasma cells within the bone marrow. The genome of the malignant plasma cells is extremely unstable characterized by a complex combination of structure and numerical abnormalities. DNA copy number variants (CNV) affects target gene expression but such affectation is not compulsory and target gene expression can be modulated. We supposed that this modulation serves as a compensatory mechanism to keep genomic homeostasis. Further compensation mechanisms exhausting leads disease aggressiveness.
Aims: The objective of our study was to define and describe influence of DNA copy number variants on gene expression level in multiple myeloma.
Material and methods: 66 newly diagnosed patients with MM were evaluated for this study. The patients' baseline characteristics were as follows: males/females: 34/32 (52% /48%); median age of 68 years (range 49-83 years). Type of M protein IgG/IgA/LC/other (total n=58); 36/11/10/1 (62%/19%/17%/2%); most of the patients had advanced stage of MM DS II+III (total n=58) n =58 (100%); ISS II+III (total n=53) n=42 (79%). CD138+ plasma cells separated by MACS. Gene expression profiling was performed using Affymetrix GeneChip Human Gene ST 1.0 array (Affymetrix). DNA copy number variations was evaluated using Agilent Human Genome CGH Microarray (4x44K), Agilent SurePrint G3 Human Genome CGH+SNP Microarray Kit (4x180K), OGT CytoSure Haematological Cancer +SNP (8x60K), Agilent SurePrint G3 Human CGH Microarray (8x60K) with proper platforms aggregation (Agilent technologies).
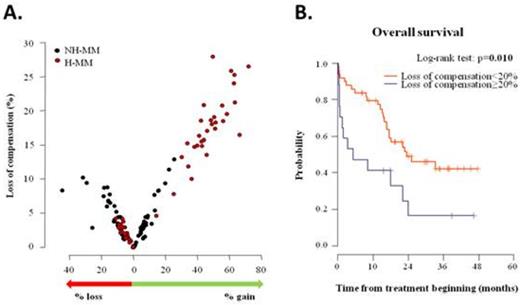
Results: Each patient had at least one CNV, the most often changes were at the level of entire chromosomes. Hyperdiploid/non-hyperdiploid patients (H-MM/NH-MM) represent 53 % (31/66) and 47 % (35/66), resp. CNV considered as uncompensated, if the value of target gene expression is lower than the 25th percentile of norm (gene expression of genes without loss or gain of DNA) for gene loss, or greater than the 75th percentile of norm for gene gain. Figure 1A shows that level of CNV modulation is determined by the number of changes that occur in a given patient. Further, ROC analysis was done to determine whether certain level of compensation is related to the overall survival and CNV compensation limit 20% (p=0.026) was established. Patients with ≥20% of decompensated CNV had significantly worse OS (survival median 5.2 month) compared to patients with <20% of decompensated CNV (survival median 23.5 month). Kaplan-Meier curves for given patients' subgroups are presented in Figure 1B.
Conclusion: Copy number variants in MM can be compensated on gene expression level. Compensatory capacity of genome is associated with total number of CNV. Patients with ≥20% of decompensated CNV had significantly worse OS.
Acknowledgment:This study was supported by grants no. MSK 02680/2014/RRC and MSK 02692/2014/RRC; MH CZ-DRO-FNOs/2014; SGS01/LF/2014-2015, SGS02/LF/2014-2015, SGS03/LF/2015-2016, NT14575, AZV 15-29508A and AZV 15-29667A
Copy number variants (CNV) modulation and their clinical impact in multiple myeloma A. The overall rate of CNV occurrence in proportion to the gene expression affectation (loss of CNV compensation) B. Overall survival in newly diagnosed MM patients with different level of CNV compensation.
Copy number variants (CNV) modulation and their clinical impact in multiple myeloma A. The overall rate of CNV occurrence in proportion to the gene expression affectation (loss of CNV compensation) B. Overall survival in newly diagnosed MM patients with different level of CNV compensation.
Hajek:Janssen-Cilag: Honoraria; Celgene, Amgen: Consultancy, Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal