Key Points
PRR deletion in T cells drastically reduces the number of peripheral and thymic CD3+ T cells.
We identify multiple stages of thymocyte development that require PRR expression.
Abstract
The (pro)renin receptor (PRR) was originally thought to be important for regulating blood pressure via the renin-angiotensin system. However, it is now emerging that PRR has instead a generic role in cellular development. Here, we have specifically deleted PRR from T cells. T-cell–specific PRR-knockout mice had a significant decrease in thymic cellularity, corresponding with a 100-fold decrease in the number of CD4+ and CD8+ thymocytes, and a large increase in double-negative (DN) precursors. Gene expression analysis on sorted DN3 thymocytes indicated that PRR-deficient thymocytes have perturbations in key cellular pathways essential at the DN3 stage, including transcription and translation. Further characterization of DN T-cell progenitors leads us to propose that PRR deletion affects thymocyte survival and development at multiple stages; from DN3 through to DN4, double-positive, and single-positive CD4 and CD8. Our study thus identifies a new role for PRR in T-cell development.
Introduction
The (pro)renin receptor (PRR) was originally identified as a receptor for (pro)renin.1 Its discovery led to an enormous amount of research in the cardiovascular field because of the involvement of (pro)renin in promoting hypertension under conditions of renin-angiotensin system hyperactivation.2 However, subsequent transgenic animal models failed to show a causal link between PRR and hypertension.2-5 Instead, it has emerged that PRR is essential for cellular development and homeostasis. PRR is ubiquitously expressed, and total knockout in mice is embryonic lethal.6 Several PRR conditional knockout (cKO) models have been characterized, all with severe phenotypes: acute kidney injury with podocyte,7,8 heart failure with cardiomyocyte,9 and impaired eye and kidney development with retinal10 and uteric bud11 cKO.
Disturbances in many processes have been observed upon deletion of PRR.5,12 These effects appear to be central to the molecular association of PRR with the vacuolar (V)- adenosine triphosphatase (ATPase), where it is proposed that PRR regulates V-ATPase activity.13 Much insight into PRR function has come from developmental biology studies, which showed that the association of PRR with the V-ATPase is essential for canonical Wnt signaling,13 an important developmental pathway. Further studies in Drosophila melanogaster identified that knockdown of PRR perturbs trafficking of Wnt signaling receptors.14,15 The role of Wnt signaling in T-cell development has been controversial, with some studies showing a role16 and others not.17,18 As PRR is highly expressed in lymphocytes,19 we hypothesized that deletion of PRR from T cells would have implications for T-cell development.
Study design
We bred ATP6AP2flox/+ females with male mice expressing Cre recombinase under the Lck promoter (supplemental Figure 1; available on the Blood Web site). Male mice with PRR conditionally deleted from T cells (ATP6AP2flox/y;LckCre) are hereafter referred to as “cKO.” Full methods are available online (see supplemental Methods).
Results and discussion
The cellularity of peripheral lymphoid organs and blood were analyzed by flow cytometry. In PRR cKO mice, a significant decrease in CD3+ T cells was observed in all tissues, with no difference in the number of B, macrophage, or dendritic cells (Figure 1A; supplemental Figures 2-3). A striking reduction in both CD4+ and CD8+ naïve T cells was evident, with no change in the number of peripheral T-cell receptor (TCR) γδ cells observed (Figure 1B). As PRR is found to be essential for homeostasis,7-11 we asked if the remaining peripheral T cells had the PRR gene excised. We performed polymerase chain reaction (PCR) that specifically amplified a segment of the PRR gene only after it was mutated by Cre recombination (supplemental Figure 4). Using this approach, we could detect the recombined gene (ie, with the PRR gene excised) in thymic but not peripheral CD90.2+ T cells, which expressed similar levels of PRR protein as control T cells (supplemental Figure 4). Together, these results indicate that peripheral cKO T cells simply do not develop or survive. Instead, those few cells that do not recombine the PRR gene have an enormous selective advantage and reach the periphery.
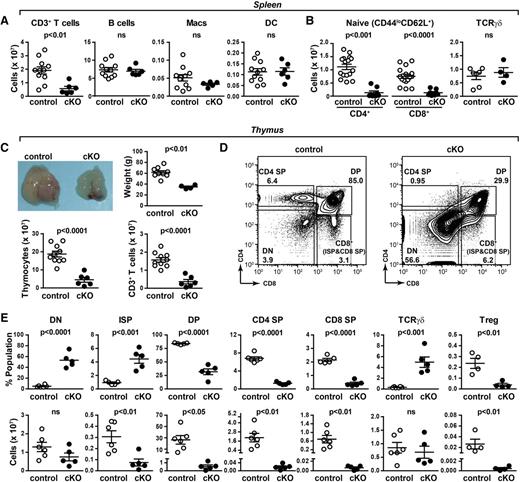
Abnormal thymic T-cell development in PRR cKO mice. (A) Absolute numbers of T (all CD3+), B, macrophage, and dendritic cells from spleens of 7- to 8-week-old mice were determined by flow cytometry. (B) Absolute numbers of naïve (CD44loCD62L+) CD4+ and CD8+ T cells and γδT cells. (C) Morphology, tissue weight, cellularity, and number of CD3+ cells of thymi from 6-week-old mice. (D) Thymocytes from 6-week-old control and cKO mice were analyzed by flow cytometry. The gating strategy for CD4 and CD8 expression is shown. (E) From panel D: the percentage (top) and number (bottom) of DN, ISP, double-positive (DP), CD4 single-positive (SP), CD8 SP, TCRγδ, and regulatory T (Treg) cells.
Abnormal thymic T-cell development in PRR cKO mice. (A) Absolute numbers of T (all CD3+), B, macrophage, and dendritic cells from spleens of 7- to 8-week-old mice were determined by flow cytometry. (B) Absolute numbers of naïve (CD44loCD62L+) CD4+ and CD8+ T cells and γδT cells. (C) Morphology, tissue weight, cellularity, and number of CD3+ cells of thymi from 6-week-old mice. (D) Thymocytes from 6-week-old control and cKO mice were analyzed by flow cytometry. The gating strategy for CD4 and CD8 expression is shown. (E) From panel D: the percentage (top) and number (bottom) of DN, ISP, double-positive (DP), CD4 single-positive (SP), CD8 SP, TCRγδ, and regulatory T (Treg) cells.
The striking reduction in peripheral naïve T cells is consistent with a defect in thymic T-cell development, so we turned our focus to this organ. A severe atrophy of PRR cKO thymi was observed, evidenced by a reduction in weight and cellularity (Figure 1C). PRR cKO thymi had a 10-fold increase in the proportion of double-negative (DN) thymocytes (Figure 1D). A significant increase in the proportion of intermediate single-positive (ISP) and TCRγδ cells was observed, whereas all other populations were decreased (Figure 1E). The number of DN cells was unchanged, whereas the numbers of ISP, DP, CD8 SP, CD4 SP, and regulatory T cells were reduced (Figure 1E). Histologic analysis indicated that the loss of SP T cells was associated with a reduction in the thymic medulla of cKO mice but preserved corticomedullary junction (supplemental Figure 5), consistent with other models of severe SP T-cell loss.20 No change in the number of TCRγδ cells was evident. This is unsurprising as the lineage commitment for TCRγδ cells begins at the DN2 stage, prior to Lck-driven Cre recombinase expression.16 Taken together, the almost 100-fold reduction in SP cells indicates that PRR cKO mice have drastically altered thymocyte development.
We next analyzed early T-cell developmental events (DN1-4). There was an increase in the proportion of DN3 cells in cKO mice, which was correlated with a decreased proportion of DN4 (Figure 2A). These proportional changes were not associated with differences in the number of DN3 or DN4 (Figure 2B). At the DN3 stage, cells commit to the T-cell lineage and rearrange the TCR β-chain gene locus. PCR products corresponding to rearranged TCRβ locus were detected in both control and cKO cells, indicating that this process was not disturbed (Figure 2C). There was no difference in intracellular TCRβ levels (Figure 2D).
PRR cKO impairs thymocyte survival and development from DN3 and beyond. (A) Representative flow cytometry of CD25 and CD44 expression in DN (CD4−CD8−) thymocytes from 39-day-old mice. (B) The frequency and actual number of DN3 and DN4 cells from panel A. (C) TCRβ gene rearrangement by PCR of sorted DN4 cells from control and cKO mice. DNA from bone marrow (BM) cells is shown as a nonrearranged control. (D) Expression of intracellular TCRβ in DN3 and DN4 thymocytes as from panel B. (E) Scatter plot showing the comparison of gene expression from microarray analysis of DN3 sorted cells from control and cKO mice. Significantly different genes (by 1-way analysis of variance [ANOVA]; q < 0.05) are labeled (black circles). For clarity, the gene 5730437N04Rik is abbreviated as “5…Rik,” and the gene 1600029D21Rik is abbreviated as “1…Rik.” Inset shows PRR expression by real-time quantitative PCR. N = 3 biological replicates. (F) GO pathways that were significantly enriched in cKO DN3 cells (false-discovery rate [FDR] <0.5) are shown. Details of the genes in each pathway are listed in supplemental Table 1. (G) Representative PCR from sorted thymocytes from 39-day-old control and cKO mice to detect the unexcised PRR gene (“floxed”) and mutated/deleted gene in cKO (“excised”). Cre recombinase expression is also shown. (H) TCRα gene rearrangement was determined in sorted DP cells from control and cKO mice by quantitative PCR with primers specific for Vα8, Vα2, and Vα10 in conjunction with different Jα primers. The P value shown for the effect of PRR deletion was calculated by 2-way ANOVA. #P < .01 by Sidak’s multiple comparison test. N = 3-4. (I) Thymocytes from control and cKO mice were cultured in vitro with interleukin 7, and the proportion of live cells was determined by flow cytometry at the desired time points. The P value shown for the effect of PRR deletion was calculated by 2-way ANOVA. **P < .01; ****P < .0001 by Sidak’s multiple comparison test. N = 5-9. (J) Schematic of T-cell development stages affected by PRR deletion.
PRR cKO impairs thymocyte survival and development from DN3 and beyond. (A) Representative flow cytometry of CD25 and CD44 expression in DN (CD4−CD8−) thymocytes from 39-day-old mice. (B) The frequency and actual number of DN3 and DN4 cells from panel A. (C) TCRβ gene rearrangement by PCR of sorted DN4 cells from control and cKO mice. DNA from bone marrow (BM) cells is shown as a nonrearranged control. (D) Expression of intracellular TCRβ in DN3 and DN4 thymocytes as from panel B. (E) Scatter plot showing the comparison of gene expression from microarray analysis of DN3 sorted cells from control and cKO mice. Significantly different genes (by 1-way analysis of variance [ANOVA]; q < 0.05) are labeled (black circles). For clarity, the gene 5730437N04Rik is abbreviated as “5…Rik,” and the gene 1600029D21Rik is abbreviated as “1…Rik.” Inset shows PRR expression by real-time quantitative PCR. N = 3 biological replicates. (F) GO pathways that were significantly enriched in cKO DN3 cells (false-discovery rate [FDR] <0.5) are shown. Details of the genes in each pathway are listed in supplemental Table 1. (G) Representative PCR from sorted thymocytes from 39-day-old control and cKO mice to detect the unexcised PRR gene (“floxed”) and mutated/deleted gene in cKO (“excised”). Cre recombinase expression is also shown. (H) TCRα gene rearrangement was determined in sorted DP cells from control and cKO mice by quantitative PCR with primers specific for Vα8, Vα2, and Vα10 in conjunction with different Jα primers. The P value shown for the effect of PRR deletion was calculated by 2-way ANOVA. #P < .01 by Sidak’s multiple comparison test. N = 3-4. (I) Thymocytes from control and cKO mice were cultured in vitro with interleukin 7, and the proportion of live cells was determined by flow cytometry at the desired time points. The P value shown for the effect of PRR deletion was calculated by 2-way ANOVA. **P < .01; ****P < .0001 by Sidak’s multiple comparison test. N = 5-9. (J) Schematic of T-cell development stages affected by PRR deletion.
We next performed gene expression analysis of sorted DN3 cells (Figure 2E; supplemental Table 1A). In cKO, 615 and 518 genes were up- and downregulated >1.5-fold. Of these, 9 genes were significantly different and encoded molecules important for processes including the cell cycle (Mcts1, Cetn2), mitochondria (Tomm7, Uqcrh) and vesicular trafficking (Ier3ip1, Rabac1). We next performed gene ontology (GO) analysis on the differentially expressed genes (Figure 2F; supplemental Table 1B). Several GO pathways were enriched in cKO DN3 cells, indicating disturbances in transcription, translation, and the mitochondria. Collectively, these represent cellular activities essential for this developmental period, providing an explanation for why PRR deletion has such a profound effect on T-cell development.
Comparing our findings to conditional deletion of the Wnt pathway in T cells, several similarities are evident: reduction in peripheral T cells and a change in the proportion of DN3:DN4.16 However, overall it is clear that PRR deletion induces a more profound phenotype. Wnt cKO led to an increase in the proportion of DN cells to 12%, and a partial block at the DN3-DN4 transition, leading to an increased number of DN3 cells and decreased DN4. In the periphery, there was a fourfold reduction in T cells, which did have the β-catenin deleted allele.16 In contrast, PRR cKO leads to an increase in the proportion of DN cells to ∼50% of thymocytes, we detect no increase in the number of DN3 cells, and we find that DN4 numbers are decreased only in old mice (data not shown). Furthermore, we are unable to detect the deleted allele in peripheral T cells, indicating that PRR-deficient T cells do not develop/survive. To understand these results further, we sorted thymocytes and again performed PCR to specifically identify cKO cells. This revealed the presence of cKO cells beyond the DN3 stage, including DN4, DP, and CD4 SP (Figure 2G). We analyzed DP cells further and observed an impaired survival and, subsequently, rearrangement of the TCR α chain (Tcra) (Figure 2H-I). Thus, given the striking severity of our phenotype, that no single precursor thymocyte population accumulates, and that we also observe defects in DP cells, we propose that deletion of PRR affects thymocyte survival and development from DN3 and beyond (Figure 2J).
Authorship
Contribution: S.G., U.M., M.G., A.M., and K.J.B. performed experiments; M.K., R.A.L., R.D., A.C., and O.D. gave advice; G.N. provided mice; and D.N.M., M.D.W., and K.J.B. designed and wrote the study.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Katrina J. Binger, Experimental and Clinical Research Center, Lindenbergerweg 80, Berlin, 13125, Germany; e-mail: katrinabinger@gmail.com; and Dominik N. Muller, Experimental and Clinical Research Center, Lindenbergerweg 80, Berlin, 13125, Germany; e-mail: dominik.mueller@mdc-berlin.de.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Jana Czychi, Ilona Kamer, Sabine Schmidt, and Dr Hans-Peter Rahn for technical assistance.
This work was supported by the National Health and Medical Research Council of Australia (K.J.B. and M.D.W.), and the German Research Foundation and German Center for Cardiovascular Research (D.N.M.).
References
Author notes
S.G. and U.M. contributed equally to this study.
M.D.W. and K.J.B. contributed equally to this study.

![Figure 2. PRR cKO impairs thymocyte survival and development from DN3 and beyond. (A) Representative flow cytometry of CD25 and CD44 expression in DN (CD4−CD8−) thymocytes from 39-day-old mice. (B) The frequency and actual number of DN3 and DN4 cells from panel A. (C) TCRβ gene rearrangement by PCR of sorted DN4 cells from control and cKO mice. DNA from bone marrow (BM) cells is shown as a nonrearranged control. (D) Expression of intracellular TCRβ in DN3 and DN4 thymocytes as from panel B. (E) Scatter plot showing the comparison of gene expression from microarray analysis of DN3 sorted cells from control and cKO mice. Significantly different genes (by 1-way analysis of variance [ANOVA]; q < 0.05) are labeled (black circles). For clarity, the gene 5730437N04Rik is abbreviated as “5…Rik,” and the gene 1600029D21Rik is abbreviated as “1…Rik.” Inset shows PRR expression by real-time quantitative PCR. N = 3 biological replicates. (F) GO pathways that were significantly enriched in cKO DN3 cells (false-discovery rate [FDR] <0.5) are shown. Details of the genes in each pathway are listed in supplemental Table 1. (G) Representative PCR from sorted thymocytes from 39-day-old control and cKO mice to detect the unexcised PRR gene (“floxed”) and mutated/deleted gene in cKO (“excised”). Cre recombinase expression is also shown. (H) TCRα gene rearrangement was determined in sorted DP cells from control and cKO mice by quantitative PCR with primers specific for Vα8, Vα2, and Vα10 in conjunction with different Jα primers. The P value shown for the effect of PRR deletion was calculated by 2-way ANOVA. #P < .01 by Sidak’s multiple comparison test. N = 3-4. (I) Thymocytes from control and cKO mice were cultured in vitro with interleukin 7, and the proportion of live cells was determined by flow cytometry at the desired time points. The P value shown for the effect of PRR deletion was calculated by 2-way ANOVA. **P < .01; ****P < .0001 by Sidak’s multiple comparison test. N = 5-9. (J) Schematic of T-cell development stages affected by PRR deletion.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/126/4/10.1182_blood-2015-03-635292/4/m_504f2.jpeg?Expires=1769111884&Signature=DROJgtkJNj2AdXSaUaBiG8obbbRCcKDOlyTfX9Sjb9cg2~caIG5YDocbvLE4Pb9h8iaDe6DZuaTaydNnzAbbvZT23F-g5Gzck5~B2po9s7trW-n1nMHpUu1ny-dB~spLi~d0nZlXBDyD80xyfBZfFTgcn53ugWvBsYgeUFUt5C92W5i7gdNNEwCqnmWWOM8MTNnDgfQMpB2E~jblZ-Xio2GYYWdwX9N9NLrXlHeD0MD-Kh5ynVMGsV9AIidpSHceH2zldSzQeAtSHTyOxwzvL6ZukxYS4hdLttQ9hka7Ak212yRrnU9XdOtZugZqhjONCRbccT0epLfyX-GoHJKIzA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal