Abstract
Since the first description of the natural history of chronic lymphocytic leukemia (CLL) by David Galton in 1966, the considerable heterogeneity in the disease course has been well recognized. The Rai and Binet staging systems described ∼40 years ago have proven to be robust prognostic tools. Over the past 2 decades, several novel biological, genetic, and molecular markers have been shown to be useful adjuncts to the Rai and Binet staging systems. In this systematic review, we examined the role of immunoglobulin heavy-chain variable region gene (IGHV) mutation status and genetic abnormalities determined by interphase fluorescence in situ hybridization (FISH) in patients with newly diagnosed CLL. The cumulative evidence presented in this systematic review is sufficient to recommend that FISH and IGHV be performed as standard clinical tests for all patients with newly diagnosed CLL in those countries with the resources to do so. In addition to clinical stage, these parameters could represent the minimal standard initial prognostic evaluation for patients with CLL. This approach will allow the application of powerful, recently developed prognostic indices (all of which are dependent on IGHV and FISH results) to all patients with newly diagnosed CLL.
Case presentation
Case 1. A 72-year-old man was incidentally discovered to have a white blood cell (WBC) count of 22 × 109/L during a routine annual visit to his primary care physician. The absolute lymphocyte count was 20 × 109/L. Peripheral blood flow cytometry revealed a clonal population of lymphocytes that coexpressed CD19, CD5, CD23, and CD20 (dim), consistent with a diagnosis of chronic lymphocytic leukemia (CLL). Serum β2-microglobulin level was normal. He was referred to a hematologist for further treatment. The patient was asymptomatic; physical examination demonstrated no organomegaly or lymphadenopathy. He was diagnosed with Rai 0 CLL.
Case 2. A 62-year-old woman was incidentally noted to have a WBC count of 15 × 109/L, with an absolute lymphocyte count of 14 × 109/L. Peripheral blood flow cytometry confirmed a diagnosis of CLL. The patient was asymptomatic; physical examination demonstrated no organomegaly or lymphadenopathy. Serum β2-microglobulin was 4.1 μg/mL (normal, 1.2-2.7 μg/mL). The patient was diagnosed with Rai 0 CLL.
What is the role of obtaining immunoglobulin heavy-chain variable region gene (IGHV) mutation status and an interphase fluorescence in situ hybridization (FISH) study at the time of CLL diagnosis in routine practice?
Background
Approximately 16 000 new cases of CLL are diagnosed each year in the United States.1 There is considerable heterogeneity in the disease course of CLL—some patients have an indolent course and live for decades without therapy, whereas others experience relatively rapid progression and succumb to the disease within a few years despite maximal therapy.2 Effective approaches to stratifying a patient’s prognosis can enable the treating physician to provide more accurate patient counseling, tailor the frequency of follow-up, and, in some cases, inform therapy selection.
The Rai and Binet staging systems were developed ∼40 years ago using readily available clinical and laboratory parameters to stratify patient risk.3,4 Despite the enduring utility of clinical staging, there remains significant clinical heterogeneity among patients within each Rai and Binet stage category. Furthermore, approximately three quarters of newly diagnosed CLL patients are diagnosed at the Rai 0/Binet A stage, where the staging systems are unable to determine the likelihood or pace of disease progression.
Although a plethora of prognostic parameters have been proposed to address this limitation over the last 35 years, IGHV mutation status and cytogenetic abnormalities identified by FISH have been the most widely studied. In 1999, 2 independent groups reported that patients with higher levels of somatic mutation in the IGHV genes of their CLL clone experienced longer progression-free survival (PFS) and overall survival (OS).5,6 Roughly 1 year after these 2 reports, Dohner and colleagues proposed a new prognostic model that categorized patients into 5 risk categories based on FISH. Using a hierarchical classification scheme, they demonstrated the shortest survival for patients with del17p13 (32 months), followed by patients with del11q23 (79 months), trisomy 12 (111 months), normal karyotype (114 months), and del13q14 as the sole abnormality (133 months).7 After these seminal observations, several studies have demonstrated the consistent and robust ability of IGHV mutation status and interphase FISH to stratify patient outcome. Here, we performed a systematic review evaluating the prognostic utility of these 2 parameters in patients with newly diagnosed CLL.
Methods
Eligibility criteria and literature search
We performed a literature search to identify studies on the prognostic value of IGHV mutation status and FISH in CLL with the aid of an experienced medical librarian. We applied no language restrictions. We searched 5 databases (Ovid MEDLINE, Ovid EMBASE, Ovid CENTRAL, Web of Science, and Scopus) to identify all citations from January 1999 to April 2015 describing the role of IGHV mutation testing and FISH in predicting prognosis for CLL. Ovid MEDLINE was used to design the strategy, using a combination of MeSH-controlled vocabulary and text words for each concept. The following terms were used to perform the search: immunoglobulin variable region, immunoglobulin heavy chain, genes, immunoglobulin, or IGHV (as text words); FISH, in situ hybridization, fluorescent; leukemia, lymphocytic, chronic, B-cell, or CLL. The results were imported into EndNote, and duplicate results were removed.
Full-length publications reporting on the prognostic value (eg, PFS and/or OS) of IGHV and/or FISH in patients with newly diagnosed CLL, and which included at least 200 patients, were included in the systematic review. Studies that included <200 patients, focused on treated patients, or did not report on PFS or OS were excluded. Preliminary results reported only in abstract form were excluded. Manuscripts that described the prognostic impact of IGHV mutation and FISH in the context of patients starting treatment on a clinical trial were not included, because ∼30% to 50% of patients with newly diagnosed CLL never require therapy and the focus of such studies is evaluating the impact of therapy on OS as opposed to the use of prognostic parameters for risk stratification.
Study evaluation
Two reviewers working independently considered the potential eligibility of each of the abstracts generated by the search strategy. Each abstract was evaluated independently for final study inclusion. For discrepancies arising in the data abstracting process, a third reviewer returned to the source document to determine the accurate information.
Data extraction
Data were extracted using a standardized form to enter study participant characteristics, proportion of patients who had IGHV mutation status and FISH testing performed, and PFS and OS for all patients. Data extraction was performed in duplicate by 2 reviewers.
Meta-analysis
We performed generic inverse variance meta-analyses using random-effects models to calculate pooled hazard ratios (HR) with 95% confidence intervals (CI) for PFS and OS from multivariable study results for both IGHV mutation status and FISH results. Heterogeneity was assessed using the I2 statistic. Statistical analyses were performed using Review Manager 5.3 (Cochrane Collaboration) software.
Results
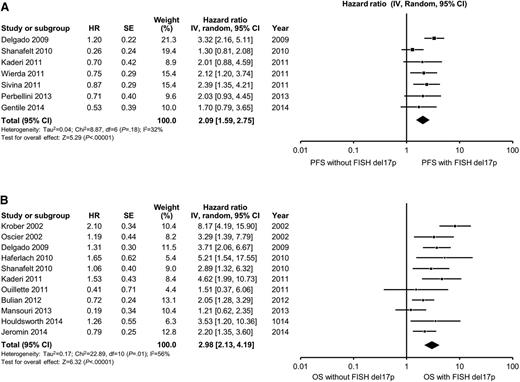
The search criteria described in “Methods” identified 1450 citations. After independent evaluation of all 1450 studies by 2 reviewers, 31 studies met the criteria for inclusion in this study (Figure 1). Of these, 7 reported outcomes for IGHV mutation status only, 2 for FISH only, and 22 for both IGHV mutation status and FISH. Table 1 shows the baseline characteristics of patients, and the median PFS and median OS for both IGHV mutation status and interphase FISH for all studies.7-35 Table 2 shows the HR for PFS and OS reported for multivariate models that included both IGHV mutation status and FISH.7-35
Median PFS and OS for IGHV and FISH of all studies included in this analysis
| No. . | Reference . | n . | Median age (y) . | Males (%) . | Rai/Binet stage (%) . | Progression-free survival (y) . | Overall survival (y) . | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IGHV median* . | FISH median* . | IGHV median* . | FISH median* . | ||||||||||||||||||
| 0 or A . | I/II or B . | III/IV or C . | Mutated . | Unmutated . | 13q– . | NL . | +12 . | 11q– . | 17p– . | Mutated . | Uunmutated . | 13q– . | NL . | +12 . | 11q– . | 17p- . | |||||
| IGHV only | |||||||||||||||||||||
| 1 | 6 | 84 | 63 | 58 | 74 | 11 | 15 | — | — | 24.5 | 9.7 | ||||||||||
| 2 | 5 | 47 | 63 | 67 | 37 | 58 | 5 | — | — | — | 9 | ||||||||||
| 3 | 8 | 307 | 52 | 65 | — | — | — | — | — | — | — | ||||||||||
| 4 | 19 | 303 | 63 | 54 | 34 | 62 | 2 | 10 y: 41% | 10 y: 9% | 10 y: 87% | 10 y: 43% | ||||||||||
| 5 | 10 | 262 | 65 | 61 | 100 | 0 | 0 | 3 y: 93% | 3 y: 66% | — | — | ||||||||||
| 6 | 11 | 337 | 64 | 60 | 71 | 29 | NR | 2 | NR | 7 | |||||||||||
| 7 | 12 | 224 | 63 | 68 | 77 | 23 | 18.9 | 2.5 | 17.9 | 9.8 | |||||||||||
| FISH only | |||||||||||||||||||||
| 8 | 7 | 325 | 62 | 61 | 19 | 60 | 20 | 7.7 | 4.1 | 2.7 | 1.1 | 0.7 | 11 | 9.3 | 9.5 | 6.6 | 2.7 | ||||
| 9 | 13 | 231 | 68 | 63 | 71 | 27 | 2 | — | — | — | — | — | — | — | — | — | — | ||||
| Both IGHV and FISH (OS only) | |||||||||||||||||||||
| 10 | 14 | 325 | 62 | 61 | 20 | 60 | 20 | — | 7.5 | — | — | — | 2.5 | 5.8 | |||||||
| 11 | 15 | 205 | 62 | 58 | 82 | 10 | 7 | 25.8 | 10 | 17.5 | 24.4 | 10.2 | 9.7 | 3.9 | |||||||
| 12 | 16 | 1948 | 60 | 68 | 21 | 56 | 23 | 5 y: 89% | 5 y: 71% | 5 y: 85% | 5 y: 87% | 5 y: 83% | 5 y: 75% | 5 y: 34% | |||||||
| Both IGHV and FISH (PFS only) | |||||||||||||||||||||
| 13 | 17 | 351 | 61 | 61 | 80 | 20 | 11.8 | 4 | 12 | 5 | 5 | 4 | 3 | ||||||||
| 14 | 18 | 930 | 59 | 61 | 87 | 13 | — | — | 5 | 4 | 4 | 3 | 1 | ||||||||
| 15 | 19 | 384 | 61 | 58 | 100 | 0 | 0 | — | 4 | — | — | — | — | 4 | |||||||
| 16 | 20 | 3490 | 65 | 63 | 77 | 23 | — | 3.5 | 12 | — | 4 | 3 | — | ||||||||
| Both IGHV and FISH (PFS and OS) | |||||||||||||||||||||
| 17 | 21 | 706 | 66 | 57 | 76 | 24 | 10.5 | 2 | 9 | 1 | NR | 8 | 20.5 | 6.8 | |||||||
| 18 | 22 | 482 | 67 | 65 | — | — | — | 9.2 | 1.5 | 12 | 6.6 | NR | 1 | 1.8 | 5 y: 88% | 5 y: 81% | NR | NR | NR | NR | 2.5 |
| 19 | 23 | 2487 | 64 | 68 | 52 | 39 | 6 | 11 | 2.8 | NR | 8.7 | 5.4 | 2.4 | 5.2 | NR | 9.7 | NR | NR | 10.9 | 8.4 | 7.6 |
| 20 | 24 | 252 | 64 | 63 | 79 | 16 | 5 | NR | 1.1 | NR | NR | 1.5 | 0.5 | 0.1 | NR | 7 | NR | NR | 7 | 7.6 | 4 |
| 21 | 25 | 255 | 61 | 64 | 95 | 5 | NR | 5 | — | — | — | 4 | 2.6 | — | 3.2 | — | — | — | 3.3 | 5 | |
| 22 | 26 | 1154 | 66 | — | 100 | 0 | 0 | NR | 5 | 22 | 3.6 | 23 | 13 | 23 | 13 | ||||||
| 23 | 27 | 620 | 65 | 60 | 48 | 44 | 8 | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 24 | 28 | 292 | 66% >60 y | 55 | 72 | 28 | 3 y: 58% | 3 y: 23% | 3 y: 49% | 3 y: 37% | 3 y: 28% | 3 y: 21% | 3 y: 24% | 10 y: 75% | 10 y: 41% | 10 y: 93% | 10 y: NR | 10 y: 72% | 10 y: 67% | 10 y: 0% | |
| 25 | 29 | 265 | 64 | 64 | 78 | 22 | NR | 1 | NR | NR | 2 | 1 | <1 | NR | 7 | NR | NR | 10 | 9 | 4 | |
| 26 | 30 | 311 | 66 | 61 | 78 | 15 | 6 | NR | 2 | 5.2 | 1.5 | — | — | — | — | — | — | — | |||
| 27 | 31 | 1274 | 66 | 55 | 80 | 20 | — | — | NR | 8 | 8 | — | — | — | — | 5 y: 84% | 5 y: 78% | 5 y: 63% | 5 y: 54% | 5 y: 48% | |
| 28 | 32 | 949 | 65 | 58 | 57 | 33 | 10 | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 29 | 33 | 468 | 53% >60 y | 59 | 100 | 0 | 0 | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 30 | 34 | 288 | — | — | 95 | 5 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | |
| 31 | 35 | 1160 | 65 | 65 | — | — | — | — | — | 9.1 | 8.4 | 4.2 | 2.3 | 3.7 | — | — | 7.5 | 7.5 | 7.5 | 5.7 | 4.2 |
| No. . | Reference . | n . | Median age (y) . | Males (%) . | Rai/Binet stage (%) . | Progression-free survival (y) . | Overall survival (y) . | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IGHV median* . | FISH median* . | IGHV median* . | FISH median* . | ||||||||||||||||||
| 0 or A . | I/II or B . | III/IV or C . | Mutated . | Unmutated . | 13q– . | NL . | +12 . | 11q– . | 17p– . | Mutated . | Uunmutated . | 13q– . | NL . | +12 . | 11q– . | 17p- . | |||||
| IGHV only | |||||||||||||||||||||
| 1 | 6 | 84 | 63 | 58 | 74 | 11 | 15 | — | — | 24.5 | 9.7 | ||||||||||
| 2 | 5 | 47 | 63 | 67 | 37 | 58 | 5 | — | — | — | 9 | ||||||||||
| 3 | 8 | 307 | 52 | 65 | — | — | — | — | — | — | — | ||||||||||
| 4 | 19 | 303 | 63 | 54 | 34 | 62 | 2 | 10 y: 41% | 10 y: 9% | 10 y: 87% | 10 y: 43% | ||||||||||
| 5 | 10 | 262 | 65 | 61 | 100 | 0 | 0 | 3 y: 93% | 3 y: 66% | — | — | ||||||||||
| 6 | 11 | 337 | 64 | 60 | 71 | 29 | NR | 2 | NR | 7 | |||||||||||
| 7 | 12 | 224 | 63 | 68 | 77 | 23 | 18.9 | 2.5 | 17.9 | 9.8 | |||||||||||
| FISH only | |||||||||||||||||||||
| 8 | 7 | 325 | 62 | 61 | 19 | 60 | 20 | 7.7 | 4.1 | 2.7 | 1.1 | 0.7 | 11 | 9.3 | 9.5 | 6.6 | 2.7 | ||||
| 9 | 13 | 231 | 68 | 63 | 71 | 27 | 2 | — | — | — | — | — | — | — | — | — | — | ||||
| Both IGHV and FISH (OS only) | |||||||||||||||||||||
| 10 | 14 | 325 | 62 | 61 | 20 | 60 | 20 | — | 7.5 | — | — | — | 2.5 | 5.8 | |||||||
| 11 | 15 | 205 | 62 | 58 | 82 | 10 | 7 | 25.8 | 10 | 17.5 | 24.4 | 10.2 | 9.7 | 3.9 | |||||||
| 12 | 16 | 1948 | 60 | 68 | 21 | 56 | 23 | 5 y: 89% | 5 y: 71% | 5 y: 85% | 5 y: 87% | 5 y: 83% | 5 y: 75% | 5 y: 34% | |||||||
| Both IGHV and FISH (PFS only) | |||||||||||||||||||||
| 13 | 17 | 351 | 61 | 61 | 80 | 20 | 11.8 | 4 | 12 | 5 | 5 | 4 | 3 | ||||||||
| 14 | 18 | 930 | 59 | 61 | 87 | 13 | — | — | 5 | 4 | 4 | 3 | 1 | ||||||||
| 15 | 19 | 384 | 61 | 58 | 100 | 0 | 0 | — | 4 | — | — | — | — | 4 | |||||||
| 16 | 20 | 3490 | 65 | 63 | 77 | 23 | — | 3.5 | 12 | — | 4 | 3 | — | ||||||||
| Both IGHV and FISH (PFS and OS) | |||||||||||||||||||||
| 17 | 21 | 706 | 66 | 57 | 76 | 24 | 10.5 | 2 | 9 | 1 | NR | 8 | 20.5 | 6.8 | |||||||
| 18 | 22 | 482 | 67 | 65 | — | — | — | 9.2 | 1.5 | 12 | 6.6 | NR | 1 | 1.8 | 5 y: 88% | 5 y: 81% | NR | NR | NR | NR | 2.5 |
| 19 | 23 | 2487 | 64 | 68 | 52 | 39 | 6 | 11 | 2.8 | NR | 8.7 | 5.4 | 2.4 | 5.2 | NR | 9.7 | NR | NR | 10.9 | 8.4 | 7.6 |
| 20 | 24 | 252 | 64 | 63 | 79 | 16 | 5 | NR | 1.1 | NR | NR | 1.5 | 0.5 | 0.1 | NR | 7 | NR | NR | 7 | 7.6 | 4 |
| 21 | 25 | 255 | 61 | 64 | 95 | 5 | NR | 5 | — | — | — | 4 | 2.6 | — | 3.2 | — | — | — | 3.3 | 5 | |
| 22 | 26 | 1154 | 66 | — | 100 | 0 | 0 | NR | 5 | 22 | 3.6 | 23 | 13 | 23 | 13 | ||||||
| 23 | 27 | 620 | 65 | 60 | 48 | 44 | 8 | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 24 | 28 | 292 | 66% >60 y | 55 | 72 | 28 | 3 y: 58% | 3 y: 23% | 3 y: 49% | 3 y: 37% | 3 y: 28% | 3 y: 21% | 3 y: 24% | 10 y: 75% | 10 y: 41% | 10 y: 93% | 10 y: NR | 10 y: 72% | 10 y: 67% | 10 y: 0% | |
| 25 | 29 | 265 | 64 | 64 | 78 | 22 | NR | 1 | NR | NR | 2 | 1 | <1 | NR | 7 | NR | NR | 10 | 9 | 4 | |
| 26 | 30 | 311 | 66 | 61 | 78 | 15 | 6 | NR | 2 | 5.2 | 1.5 | — | — | — | — | — | — | — | |||
| 27 | 31 | 1274 | 66 | 55 | 80 | 20 | — | — | NR | 8 | 8 | — | — | — | — | 5 y: 84% | 5 y: 78% | 5 y: 63% | 5 y: 54% | 5 y: 48% | |
| 28 | 32 | 949 | 65 | 58 | 57 | 33 | 10 | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 29 | 33 | 468 | 53% >60 y | 59 | 100 | 0 | 0 | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| 30 | 34 | 288 | — | — | 95 | 5 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | |
| 31 | 35 | 1160 | 65 | 65 | — | — | — | — | — | 9.1 | 8.4 | 4.2 | 2.3 | 3.7 | — | — | 7.5 | 7.5 | 7.5 | 5.7 | 4.2 |
FISH, fluorescence in situ hybridization; IGHV: immunoglobulin heavy-chain variable region gene mutation status; NR, not reached; OS, overall survival; PFS: progression-free survival.
In studies where the median was not reported, outcomes at specific time points (e.g., 3 y PFS or 10 y OS, when available) were included
Multivariable analysis results of studies included in this analysis
| No. . | Reference . | n . | Variables included in multivariable models . | HR for PFS . | HR for OS . | Other variables significant on multivariable analysis . | |||
|---|---|---|---|---|---|---|---|---|---|
| Unmutated IGHV . | High-risk FISH . | Unmutated IGHV . | High-risk FISH . | PFS . | OS . | ||||
| IGHV only | |||||||||
| 1 | 6 | 84 | — | — | — | — | — | ||
| 2 | 5 | 47 | — | — | — | — | — | ||
| 3 | 8 | 307 | ZAP-70, IGHV | 2.5 | — | Zap-70, IGHV | — | ||
| 4 | 9 | 303 | Age, Rai stage, CD49d, ZAP-70, IGHV | — | 6.5 | Rai stage, β2M, CD49d, IGHV | Age, CD49d, IGHV | ||
| 5 | 10 | 262 | IGHV, ZAP-70, CD38 | 2.5 | — | IGHV, ZAP-70, CD38 | — | ||
| 6 | 11 | 337 | — | — | — | — | — | ||
| 7 | 12 | 224 | — | — | — | — | — | ||
| FISH only | |||||||||
| 8 | 7 | 325 | LDH, WBC, Binet stage, FISH | — | 2.9 (del11q) | — | LDH, WBC, Binet stage, FISH | ||
| 8.1 (del17p) | |||||||||
| 9 | 13 | 231 | β2M, morphology, FISH | 4.7 (del11q) | — | β2M, morphology, FISH | — | ||
| Both IGHV and FISH (OS only) | |||||||||
| 10 | 14 | 325 | CD38, Age, IGHV, FISH, LDH, WBC count | 2.9 | 1.8 (del11q) | IGHV, FISH, age, LDH, WBC | |||
| 8.2 (del17p) | |||||||||
| 11 | 15 | 205 | Stage, IGHV, FISH, Sex, CD38, morphology | 6.9 | 2.0 (del11q) | Stage, IGHV, FISH | |||
| 3.3 (del17p) | |||||||||
| 12 | 16 | 1948 | Age, sex, stage, ECOG performance status, CBC, lymphocyte count, LDH, TK, β2M, ZAP-70, CD38, IGHV, FISH | 1.9 | 1.4 (del11q) | Age, sex, ECOG performance status, β2M, TK, IGHV, FISH | |||
| 6.0 (del17p) | |||||||||
| Both IGHV and FISH (PFS only) | |||||||||
| 13 | 17 | 351 | CCL3, stage, CD38, IGHV, FISH | NS | 2.4 (del11q) | CCL3, stage, CD38, FISH | |||
| 2.4 (del17p) | |||||||||
| 14 | 18 | 930 | LDH, LN number and size, IGHV, FISH | 10.7 | 1.9 (del11q) | LDH, LN number and size, IGHV, FISH | |||
| 2.1 (del17p) | |||||||||
| 15 | 19 | 384 | CLL vs MBL, IGHV, NOTCH1, FISH | 3.9 | 1.5 (del11q) | CLL, IGHV | |||
| 16 | 20 | 3490 | NOTCH1, SF3B1, TP53, IGHV, FISH | 3.7 | 1.4 (del11q) | IGHV, SF3B1, TP53 | |||
| Both IGHV and FISH (PFS and OS) | |||||||||
| 17 | 21 | 706 | Age, CD38, ZAP70, FISH, IGHV, β2M and GFR-adjusted β2M | NS | 3.3 (del11q or del17p) | NS | 3.7 (del17p or del11q) | FISH, GFR-adjusted β2M | Age, FISH, GFR-adjusted β2M |
| 18 | 22 | 482 | Age, WBC, IGHV, karyotype, FISH, t(14;18) | 4.8 | NS | NS | 5.2 (del17p) | WBC, IGHV | Age, WBC, t(14;18) |
| 19 | 23 | 2487 | Rai stage, IGHV, ALC, CD38, ZAP-70, FISH | 2.8 | 1.3 (del11q or del17p) | 2.8 | 2.9 (del11q or del17p) | Stage, ALC, CD38, IGHV | CD38, FISH |
| 20 | 24 | 252 | LPL expression, age, gender, Binet stage, IGHV, FISH, CD38 | 2.4 | 1.4 (del11q) | 1.9 | 0.9 (del11q) | CD38, IGHV | Age, stage, IGHV, FISH, LPL expression |
| 2.0 (del17p) | 4.6 (del17p) | ||||||||
| 21 | 25 | 255 | SNP, IGHV, FISH | — | — | 1.8 | 1.5 (del11q or del17p) | — | SNP |
| 22 | 26 | 1154 | CD38, age, LDT, IGHV, FISH, ZAP-70 | 3.3 | NS | 2.7 | NS | Age, LDT, CD38, IGHV | |
| 23 | 27 | 620 | Age, sex, ALC, β2M, stage, CD38, lymph node regions, ZAP-70, year of diagnosis, therapy | 3.1 | 2.0 (del11q) | 2.0 | 2.1 (del17p) | β2M, IGHV, FISH | Age, sex, β2M, stage, IGHV, FISH |
| 24 | 28 | 292 | LDH, sex, β2M, IL-10 SNP, IGHV | 2.0 | — | 2.1 | — | Sex, LDH, β2M, IL-10 SNP, IGHV | Stage, IL-10 SNP, IGHV |
| 25 | 29 | 265 | Age, Binet Stage, Sex, CD38, FISH, telomere length, TP53/NOTCH1/SF3B1 mutation | — | 2.1 (del11q) | — | 1.2 (del11q) | CD38, genetic profile, telomere length, FISH | Age, stage, CD38, genetic profile, telomere length |
| 26 | 30 | 311 | LAIR1 expression, IGHV, Binet stage, CD49d | 2.7 | 2.0 (del11q or del17p) | — | — | LAIR1, stage, IGHV | — |
| 27 | 31 | 1274 | age, stage, integrated cytogenetic and mutational model, IGHV | — | — | 1.6 | — | — | Age, stage, IGHV, integrated cytogenetic and mutational model |
| 28 | 32 | 949 | Stage, β2M, FISH, ZAP-70, IGHV, NOTCH1, SF3B1, CIRS | NS | NS | — | NS | Stage, β2M, ZAP-70 | Stage, β2M, ZAP-70, CIRS |
| 29 | 33 | 468 | Rai stage, ALC, β2M, CD38, gene profile, IGHV, FISH | 2.6 | 1.7 (del11q or del17p) | — | — | Rai stage, ALC, β2M, progression-risk score | — |
| 30 | 34 | 288 | Array CGH, IGHV | 5.6 | - | 6.6 | 3.5 (del17p)* | Array CGH, IGHV | |
| 31 | 35 | 1160 | SF3B1, NOTCH1, IGHV, TP53, FISH, sex, age, CD38, ZAP-70 | 3.0 | 1.5 (del11q) | 2.2 | 2.2 (del17p)† | SF3B1, IGHV, FISH | SF3B1, TP53, age, CD38 |
| No. . | Reference . | n . | Variables included in multivariable models . | HR for PFS . | HR for OS . | Other variables significant on multivariable analysis . | |||
|---|---|---|---|---|---|---|---|---|---|
| Unmutated IGHV . | High-risk FISH . | Unmutated IGHV . | High-risk FISH . | PFS . | OS . | ||||
| IGHV only | |||||||||
| 1 | 6 | 84 | — | — | — | — | — | ||
| 2 | 5 | 47 | — | — | — | — | — | ||
| 3 | 8 | 307 | ZAP-70, IGHV | 2.5 | — | Zap-70, IGHV | — | ||
| 4 | 9 | 303 | Age, Rai stage, CD49d, ZAP-70, IGHV | — | 6.5 | Rai stage, β2M, CD49d, IGHV | Age, CD49d, IGHV | ||
| 5 | 10 | 262 | IGHV, ZAP-70, CD38 | 2.5 | — | IGHV, ZAP-70, CD38 | — | ||
| 6 | 11 | 337 | — | — | — | — | — | ||
| 7 | 12 | 224 | — | — | — | — | — | ||
| FISH only | |||||||||
| 8 | 7 | 325 | LDH, WBC, Binet stage, FISH | — | 2.9 (del11q) | — | LDH, WBC, Binet stage, FISH | ||
| 8.1 (del17p) | |||||||||
| 9 | 13 | 231 | β2M, morphology, FISH | 4.7 (del11q) | — | β2M, morphology, FISH | — | ||
| Both IGHV and FISH (OS only) | |||||||||
| 10 | 14 | 325 | CD38, Age, IGHV, FISH, LDH, WBC count | 2.9 | 1.8 (del11q) | IGHV, FISH, age, LDH, WBC | |||
| 8.2 (del17p) | |||||||||
| 11 | 15 | 205 | Stage, IGHV, FISH, Sex, CD38, morphology | 6.9 | 2.0 (del11q) | Stage, IGHV, FISH | |||
| 3.3 (del17p) | |||||||||
| 12 | 16 | 1948 | Age, sex, stage, ECOG performance status, CBC, lymphocyte count, LDH, TK, β2M, ZAP-70, CD38, IGHV, FISH | 1.9 | 1.4 (del11q) | Age, sex, ECOG performance status, β2M, TK, IGHV, FISH | |||
| 6.0 (del17p) | |||||||||
| Both IGHV and FISH (PFS only) | |||||||||
| 13 | 17 | 351 | CCL3, stage, CD38, IGHV, FISH | NS | 2.4 (del11q) | CCL3, stage, CD38, FISH | |||
| 2.4 (del17p) | |||||||||
| 14 | 18 | 930 | LDH, LN number and size, IGHV, FISH | 10.7 | 1.9 (del11q) | LDH, LN number and size, IGHV, FISH | |||
| 2.1 (del17p) | |||||||||
| 15 | 19 | 384 | CLL vs MBL, IGHV, NOTCH1, FISH | 3.9 | 1.5 (del11q) | CLL, IGHV | |||
| 16 | 20 | 3490 | NOTCH1, SF3B1, TP53, IGHV, FISH | 3.7 | 1.4 (del11q) | IGHV, SF3B1, TP53 | |||
| Both IGHV and FISH (PFS and OS) | |||||||||
| 17 | 21 | 706 | Age, CD38, ZAP70, FISH, IGHV, β2M and GFR-adjusted β2M | NS | 3.3 (del11q or del17p) | NS | 3.7 (del17p or del11q) | FISH, GFR-adjusted β2M | Age, FISH, GFR-adjusted β2M |
| 18 | 22 | 482 | Age, WBC, IGHV, karyotype, FISH, t(14;18) | 4.8 | NS | NS | 5.2 (del17p) | WBC, IGHV | Age, WBC, t(14;18) |
| 19 | 23 | 2487 | Rai stage, IGHV, ALC, CD38, ZAP-70, FISH | 2.8 | 1.3 (del11q or del17p) | 2.8 | 2.9 (del11q or del17p) | Stage, ALC, CD38, IGHV | CD38, FISH |
| 20 | 24 | 252 | LPL expression, age, gender, Binet stage, IGHV, FISH, CD38 | 2.4 | 1.4 (del11q) | 1.9 | 0.9 (del11q) | CD38, IGHV | Age, stage, IGHV, FISH, LPL expression |
| 2.0 (del17p) | 4.6 (del17p) | ||||||||
| 21 | 25 | 255 | SNP, IGHV, FISH | — | — | 1.8 | 1.5 (del11q or del17p) | — | SNP |
| 22 | 26 | 1154 | CD38, age, LDT, IGHV, FISH, ZAP-70 | 3.3 | NS | 2.7 | NS | Age, LDT, CD38, IGHV | |
| 23 | 27 | 620 | Age, sex, ALC, β2M, stage, CD38, lymph node regions, ZAP-70, year of diagnosis, therapy | 3.1 | 2.0 (del11q) | 2.0 | 2.1 (del17p) | β2M, IGHV, FISH | Age, sex, β2M, stage, IGHV, FISH |
| 24 | 28 | 292 | LDH, sex, β2M, IL-10 SNP, IGHV | 2.0 | — | 2.1 | — | Sex, LDH, β2M, IL-10 SNP, IGHV | Stage, IL-10 SNP, IGHV |
| 25 | 29 | 265 | Age, Binet Stage, Sex, CD38, FISH, telomere length, TP53/NOTCH1/SF3B1 mutation | — | 2.1 (del11q) | — | 1.2 (del11q) | CD38, genetic profile, telomere length, FISH | Age, stage, CD38, genetic profile, telomere length |
| 26 | 30 | 311 | LAIR1 expression, IGHV, Binet stage, CD49d | 2.7 | 2.0 (del11q or del17p) | — | — | LAIR1, stage, IGHV | — |
| 27 | 31 | 1274 | age, stage, integrated cytogenetic and mutational model, IGHV | — | — | 1.6 | — | — | Age, stage, IGHV, integrated cytogenetic and mutational model |
| 28 | 32 | 949 | Stage, β2M, FISH, ZAP-70, IGHV, NOTCH1, SF3B1, CIRS | NS | NS | — | NS | Stage, β2M, ZAP-70 | Stage, β2M, ZAP-70, CIRS |
| 29 | 33 | 468 | Rai stage, ALC, β2M, CD38, gene profile, IGHV, FISH | 2.6 | 1.7 (del11q or del17p) | — | — | Rai stage, ALC, β2M, progression-risk score | — |
| 30 | 34 | 288 | Array CGH, IGHV | 5.6 | - | 6.6 | 3.5 (del17p)* | Array CGH, IGHV | |
| 31 | 35 | 1160 | SF3B1, NOTCH1, IGHV, TP53, FISH, sex, age, CD38, ZAP-70 | 3.0 | 1.5 (del11q) | 2.2 | 2.2 (del17p)† | SF3B1, IGHV, FISH | SF3B1, TP53, age, CD38 |
β2M, serum β2-microglobulin; CCL3, chemokine ligand 3; CGH, comparative genomic hybridization; ECOG, Eastern Cooperative Oncology Group; GFR, glomerular filtration rate; FISH, fluorescence in situ hybridization; HR, hazard ratio; IGHV, immunoglobulin heavy-chain variable region gene mutation status; LAIR1, leukocyte-associated immunoglobulin-like receptor 1; LDH, lactate dehydrogenase; LPL, lipoprotein lipase; MBL, monoclonal B-cell lymphocytosis; OS, overall survival; PFS, progression-free survival; SNP, single-nucleotide polymorphism; TK, serum thymidine kinase; WBC, white cell count.
del17p identified by array CGH also.
TP53 mutation included with del17p.
IGHV mutation status
The median PFS for patients with unmutated IGHV genes (range, 1-5 years) was significantly shorter than for those with mutated IGHV genes (range, 9.2-18.9 years) across all studies. Similarly, the median OS for patients with unmutated IGHV (range, 3.2-10 years) was also significantly shorter than for those with mutated IGHV (range, 17.9-25.8 years) across all studies.
Interphase FISH
The median PFS of patients with high-risk FISH (including del17p13 and del11q23; range, 0.1-5.2 years) was significantly shorter than those with low/intermediate-risk FISH (including del13q, normal, and trisomy 12; range, 1.5-22 years). The median OS of patients with high-risk FISH (range, 3.3-9.7 years) was also significantly shorter than those with low/intermediate-risk FISH (range, 7.5-20.5 years).
Meta-analyses
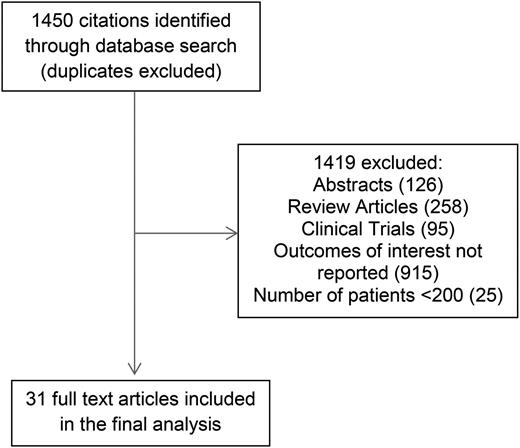
In multivariable analyses, the HR for unmutated IGHV ranged from 2.0 to 10.7 for PFS and 1.6 to 6.9 for OS compared with mutated IGHV. IGHV remained an independent predictor of PFS in 15 of 18 studies reporting the results of multivariable analysis, including 12 of 15 studies adjusting for the prognostic impact of FISH. The pooled HR for PFS was 3.2 (95% CI, 2.8-3.7; P < .0001; I2 = 20%; Figure 2A). With respect to OS, IGHV remained an independent predictor of OS in 11 of 15 studies reporting the results of multivariable analysis, including 10 of 14 studies adjusting for the prognostic impact of FISH. The pooled HR for OS was 2.4 (95% CI, 2.0-3.0; P < .0001; I2 = 50%; Figure 2B). Heterogeneity was not explained by differing inclusion criteria (such as disease stage) across studies, and summary HR estimates were similar across study classes.
Meta-analysis of studies according to IGHV mutation status. Forest plot of studies reporting (A) progression-free survival (PFS) according to IGHV mutation status, and (B) overall survival (OS) according to IGHV mutation status. df, degree of freedom; IV, inverse variance; Random, random-effects model; SE, standard error; Z, Z value.
Meta-analysis of studies according to IGHV mutation status. Forest plot of studies reporting (A) progression-free survival (PFS) according to IGHV mutation status, and (B) overall survival (OS) according to IGHV mutation status. df, degree of freedom; IV, inverse variance; Random, random-effects model; SE, standard error; Z, Z value.
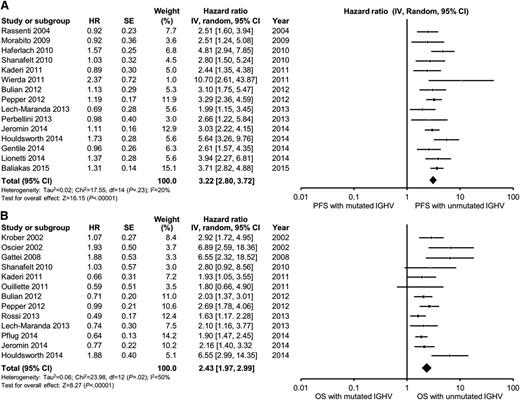
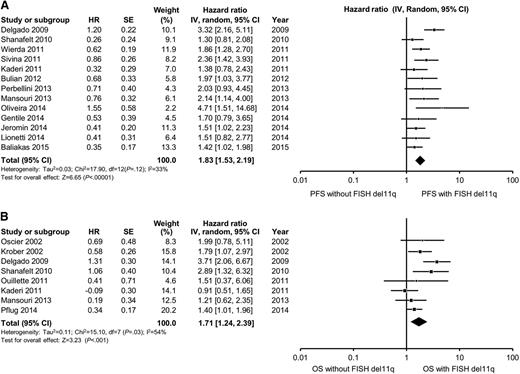
In multivariable analyses, the hazard ratio for high-risk FISH (defined as the presence of either del11q23 or del17p13) ranged from 1.3 to 4.7 for PFS and from 0.9 to 8.2 for OS. High-risk FISH remained an independent predictor of PFS in 8 of 17 studies reporting the results of multivariable analysis, including in 6 of 15 studies adjusting for the prognostic impact of IGHV. The pooled HR for PFS for studies reporting del11q23 FISH was 1.8 (95% CI, 1.5-2.2; P < .0001; I2 = 33%; Figure 3A). The pooled HR for PFS for studies reporting del17p13 FISH was 2.1 (95% CI, 1.6-2.7; P < .0001; I2 = 32%; Figure 4A). With respect to OS, FISH remained an independent predictor of OS in 10 of 14 studies reporting the results of multivariable analysis, including in 10 of 13 studies adjusting for the prognostic impact of IGHV. The pooled HR for OS for studies reporting del11q23 FISH was 1.7 (95% CI, 1.2-3.3; P = .001; I2 = 54%; Figure 3B). The pooled HR for OS for studies reporting del17p13 FISH was 3.0 (95% CI, 2.1-4.2; P < .0001; I2 = 56%; Figure 4B). Heterogeneity was not explained by differing inclusion criteria across studies or specific high-risk FISH definitions, and summary HR estimates were similar across study classes.
Meta-analysis of studies according to del11q23 status by FISH. Forest plot of studies reporting (A) progression-free survival (PFS) according to del11q23 status by FISH, and (B) overall survival (OS) according to del11q23 status by FISH. df, degree of freedom; IV, inverse variance; Random, random-effects model; SE, standard error; Z, Z value. Dohner and collegues7 and Krober and collegues17 report on the influence of FISH on OS among the same group of patients (n = 325). For the purposes of this meta-analysis, we included data reported by Krober and colleagues only.
Meta-analysis of studies according to del11q23 status by FISH. Forest plot of studies reporting (A) progression-free survival (PFS) according to del11q23 status by FISH, and (B) overall survival (OS) according to del11q23 status by FISH. df, degree of freedom; IV, inverse variance; Random, random-effects model; SE, standard error; Z, Z value. Dohner and collegues7 and Krober and collegues17 report on the influence of FISH on OS among the same group of patients (n = 325). For the purposes of this meta-analysis, we included data reported by Krober and colleagues only.
Meta-analysis of studies according to del17p13 status by FISH. Forest plot of studies reporting (A) progression-free survival (PFS) according to del17p13 status by FISH, and (B) overall survival (OS) according to del17p13 status by FISH. df, degree of freedom; IV, inverse variance; Random, random-effects model; SE, standard error; Z, Z value. Dohner and collegues7 and Krober and collegues17 report on the influence of FISH on OS among the same group of patients (n = 325). For the purposes of this meta-analysis, we included data reported by Krober and colleagues only.
Meta-analysis of studies according to del17p13 status by FISH. Forest plot of studies reporting (A) progression-free survival (PFS) according to del17p13 status by FISH, and (B) overall survival (OS) according to del17p13 status by FISH. df, degree of freedom; IV, inverse variance; Random, random-effects model; SE, standard error; Z, Z value. Dohner and collegues7 and Krober and collegues17 report on the influence of FISH on OS among the same group of patients (n = 325). For the purposes of this meta-analysis, we included data reported by Krober and colleagues only.
Discussion
The clinical course of individuals with early-stage CLL is highly variable and difficult to predict. Although the Rai/Binet staging classifications have been the international gold standards for CLL prognostication for the last 40 years, both staging systems lack the ability to predict outcomes for individual patients.3,4 Several additional novel prognostic parameters, such as sequencing for recurrent genetic abnormalities, have been identified over the last decade.36 When determining the role for these new markers, it is critical to assess their incremental value relative to existing prognostic tools. Thus, consensus on the standard prognostic evaluation is necessary to define the platform that these new tests aim to improve upon.
IGHV and FISH were first reported as prognostic parameters ∼15 to 17 years ago. Nonetheless, at the time of the last consensus guidelines reported in 2008, they were not recommended as standard tests in the routine care of patients with CLL.37 It is notable that ∼90% of patients included in this analysis were reported after the 2008 International Workshop on Chronic Lymphocytic Leukemia (IWCLL) guidelines were released. Several recent efforts have attempted to develop comprehensive approaches incorporating clinical, serum, genetic, and molecular markers with independent prognostic value into a single risk score for patients with CLL (eg, CLL International Prognostic Index, German CLL Index, MD Anderson Prognosis Score).16,18,38 It should be noted that IGHV and FISH were selected for inclusion in all of these models based on the incremental and independent prognostic information they provide.
There are several caveats to the routine use of these tests in standard clinical practice. Although the results of this analysis suggest that both IGHV mutation testing and FISH results are powerful prognostic tests for all patients with CLL, they should not be used to initiate CLL-specific therapy. Only patients with CLL who meet indication for therapy based on the 2008 IWCLL guidelines37 should receive treatment, regardless of the information obtained by prognostic testing. The only exception to this would be in the context of a clinical trial where an early intervention strategy is being used for patients at “high-risk” for CLL who do not meet the traditional indications for therapy, as was done in the recently reported CLL12 trial.39 In addition, the median age of patients included in this analysis was 64 years, which is younger than an average patient with CLL seen in practice (∼72 years). It is unclear whether IGHV mutation and FISH are equally powerful prognostic markers in these older individuals with CLL. Finally, it should be noted that the treatment landscape for CLL has dramatically improved with the approval of novel signal inhibitors, such as ibrutinib40 and idelalisib.41 Although these treatments may improve the OS of patients with CLL, they will not influence the utility of IGHV and FISH testing for predicting time to first therapy in patients with newly diagnosed CLL.
Collectively, the results of this systematic review illustrate the robust and consistent prognostic value of both IGHV and FISH independent of clinical stage in patients with newly diagnosed and/or previously untreated CLL. The bulk of the evidence also indicates that IGHV and FISH provide complementary information with respect to both PFS and OS. A greater understanding of the risk of disease progression at the time of CLL diagnosis can help (1) counsel patients appropriately; (2) define the appropriate follow-up interval (shorter interval for high-risk patients); and (3) potentially treat high-risk patients on early intervention protocols.
Recommendations
Based on the experience summarized in this review, we believe the evidence is sufficient to recommend that FISH and IGHV be recommended as standard clinical tests for all patients with newly diagnosed CLL in those countries with the resources to do so. This change will help define the minimal standard initial prognostic evaluation for patients with CLL and help facilitate use of the powerful, recently developed, integrated prognostic indices,16,18,38 all of which are dependent on these 2 variables.
Resolution of cases
Case 1
The patient underwent additional testing for IGHV gene mutation and interphase FISH. The CLL B cells showed mutated IGHV, and FISH demonstrated loss in the long arm of chromosome 13 (del13q). When we considered the patient’s age, clinical stage, and β2-microglobulin level, he was deemed to have minimal risk disease as categorized by the recently devised CLL International Prognostic Index,38 with ∼95% 5-year life expectancy and only ∼20% likelihood of requiring treatment within the following 5 years. The patient was advised to have an annual follow-up visit with his hematologist to assess for disease progression. Supportive care measures including age-appropriate cancer screening (including annual whole-body skin examination) and appropriate vaccinations for immunocompromised patients were also recommended.42
Case 2
The patient underwent additional testing for IGHV gene mutation and interphase FISH. The CLL B-cells showed an unmutated IGHV and FISH demonstrated a loss in the short arm of chromosome 17 (del17p13). When considered with the patient’s age, clinical stage, and β2-microglobulin level, she was considered to have high risk of progressive disease as categorized by the recently devised CLL International Prognostic Index, with ∼25% life expectancy and a very high likelihood of requiring treatment within the following 12 months. Although the patient was classified as having high-risk disease, she did not meet the 2008 IWCLL guidelines for starting therapy, and was therefore advised to follow-up with a hematologist every 3 months for the first year to assess for disease progression. In addition to the supportive care measures outlined for the patient in Case 1, the patient underwent human leukocyte antigen typing.
Acknowledgments
The authors acknowledge the assistance of Patricia J. Erwin, senior librarian and Assistant Professor of Medical Education at Mayo Medical School, for her assistance in identifying relevant citations from the literature for inclusion in this manuscript.
Dr Shanafelt is a clinical scholar of the Leukemia and Lymphoma Society.
Authorship
Contribution: S.A.P., P.S., M.T., C.P.W., and T.D.S. designed research and analyzed data; S.A.P., P.S., and M.T. performed research; and S.A.P. and T.D.S. wrote the paper.
Conflict-of-interest disclosure: S.A.P. has participated in advisory boards and received research support from Pharmacyclics. He was not personally compensated for the advisory board or the research support. T.D.S. has received research support from Genentech, Glaxo-Smith-Kline, Cephalon, Hospira, Celgene, Janssen, and Polyphenon E International and Pharmacyclics. He was not personally compensated for the research support. The remaining authors declare no competing financial interests.
Correspondence: Tait D. Shanafelt, Division of Hematology, Department of Medicine, Mayo Clinic, 200 First St SW, Rochester, MN 55905; e-mail: shanafelt.tait@mayo.edu.