Abstract
Until recently our approach to analyzing human genetic diseases has been to accurately phenotype patients and sequence the genes known to be associated with those phenotypes; for example, in thalassemia, the globin loci are analyzed. Sequencing has become increasingly accessible, and thus a larger panel of genes can be analyzed and whole exome and/or whole genome sequencing can be used when no variants are found in the candidate genes. By using such approaches in patients with unexplained anemias, we have discovered that a broad range of hitherto unrelated human red cell disorders are caused by variants in KLF1, a master regulator of erythropoiesis, which were previously considered to be extremely rare causes of human genetic disease.
Introduction
Over the past 5 years, more than 20 reports have identified large numbers of individuals carrying KLF1 variants. Their hematologic phenotypes range from the clinically unremarkable inhibitor of Lutheran (In(Lu)) type of the Lu(a-b-) blood group,1 to a mild increase in the level of fetal hemoglobin (HbF; α2γ2),2 to severe dyserythropoietic anemia3-5 and, in the most extreme cases, hydrops fetalis secondary to profound anemia.6 We now know that some KLF1 variants reach polymorphic frequencies in populations in which hemoglobinopathies are commonly found.7,8 This suggests that KLF1 variants have been under selection and, like the hemoglobinopathies, afford some degree of protection against malaria.9 The surprise has been discovering how common KLF1 variants are and how diverse their phenotypes are: since 2010, more than 65 different variants have been described. These observations suggest that many currently unexplained but loosely grouped human genetic diseases arise from variants in master regulators of gene expression in the affected organ systems. Here we use KLF1 as an example and discuss how extensive DNA sequencing may be used to improve health in humans.
KLF1 is a master regulator of erythropoiesis
KLF1 was discovered in 199210 ; its original name (erythroid Krüppel-like factor or EKLF) was coined because of its restricted expression in erythroid cells and its similarity to the pattern-determining protein Krüppel found in the fruit fly. Seventeen related Krüppel-like factors were subsequently identified, and the nomenclature was changed to reflect their order of discovery; thus, EKLF became KLF1. Inactivation of the Klf1 gene in mice showed that it is essential for erythropoiesis and activation of adult β-globin expression.11,12 The lethality of Klf1-null mutations in mouse fetuses was initially attributed to severe β-thalassemia, but this phenotype was not rescued when globin chain imbalance was corrected.13 This indicated that other essential erythroid genes are also regulated by KLF1, and it was confirmed by transcriptome analyses of Klf1-null erythroid cells.14-19 KLF1 activates genes that encode globins, heme synthesis enzymes, globin chaperones, structural membrane and cytoskeleton proteins, ion and water channels, metabolic and antioxidant enzymes, and cell cycle regulators20,21 (Figure 1). KLF1 also modulates expression of other transcription factors that work together in regulatory networks to control gene expression in erythroid cells.22-25
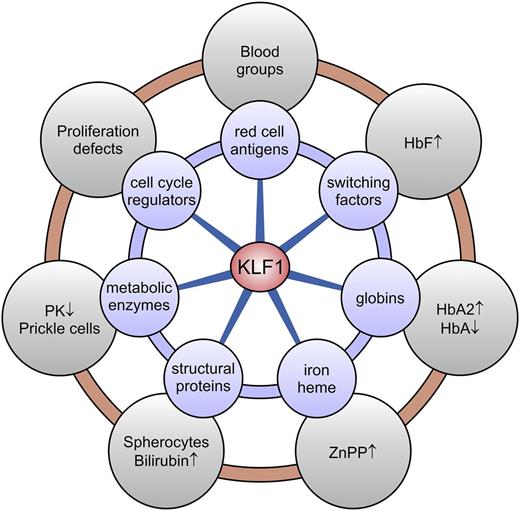
KLF1 target genes and associated clinical phenotypes. KLF1 is a master regulator of ∼700 genes in human erythroid cells involved in a wide variety of molecular processes (blue circles). Deregulated expression of a subset or all of these genes, depending on the KLF1 variant, leads to a diverse array of erythroid phenotypes (gray circles). HbA, adult hemoglobin (α2β2); HbA2, adult hemoglobin 2 (α2δ2); PK, pyruvate kinase; ZnPP, zinc protoporphyrin.
KLF1 target genes and associated clinical phenotypes. KLF1 is a master regulator of ∼700 genes in human erythroid cells involved in a wide variety of molecular processes (blue circles). Deregulated expression of a subset or all of these genes, depending on the KLF1 variant, leads to a diverse array of erythroid phenotypes (gray circles). HbA, adult hemoglobin (α2β2); HbA2, adult hemoglobin 2 (α2δ2); PK, pyruvate kinase; ZnPP, zinc protoporphyrin.
Functional domains of KLF1
KLF1 contains two short N-terminal transactivation domains (TAD1 and TAD2) with sequence similarities to TADs in other transcription factors.26,27 At the C terminus, there are 3 zinc finger domains (ZF1, ZF2, and ZF3) that enable KLF1 to bind DNA at specific sites in the genome (Figure 2). Recent studies have elucidated how the activity of KLF1 may be regulated. Throughout erythropoiesis, a significant proportion of KLF1 is found in the cytoplasm.28,29 In the mouse, a KLF1-interacting protein called friend of EKLF (FOE) may dynamically regulate retention of KLF1 in the cytoplasm via phosphorylation at serine 68.30 However, this serine is not conserved in human KLF1, and it is therefore likely that additional protein modifications or alternate mechanisms are involved in nuclear-cytoplasmic shuttling. When in the nucleus, KLF1 recruits histone modifiers (p300 and CBP),26,31 the H3.3 chaperone HIRA,32 and chromatin remodelers33,34 to specific regulatory elements and controls gene expression by working primarily as a transcriptional activator,18 although it may also repress some genes.19,35-37 As a result, KLF1 plays a critical role in establishing the correct epigenetic landscape at target gene loci in addition to its direct role in transcription.
Functional domains of KLF1 and variants reported in the literature. The KLF1 protein (362 amino acids) contains two N-terminal transactivation domains (TAD1 and TAD2) which are required for it to work as a transcriptional activator. The 3 zinc fingers (ZF1, ZF2, and ZF3) located at the C terminus form the DNA-binding domain, which enables KLF1 to bind to specific sites in the genome, typically CACCC boxes and related GC-rich elements. Residues conserved in all human KLF factors are indicated. The cysteine and histidine residues involved in zinc coordination are highlighted in blue; residues contacting DNA are highlighted in yellow. The arrows point to residues making base-specific contacts thus recognizing KLF1-binding sites in the genome. Variants are color-coded: class 1, green; class 2, blue; class 3, red; class 4, black. See supplemental Table 1 for details and references.
Functional domains of KLF1 and variants reported in the literature. The KLF1 protein (362 amino acids) contains two N-terminal transactivation domains (TAD1 and TAD2) which are required for it to work as a transcriptional activator. The 3 zinc fingers (ZF1, ZF2, and ZF3) located at the C terminus form the DNA-binding domain, which enables KLF1 to bind to specific sites in the genome, typically CACCC boxes and related GC-rich elements. Residues conserved in all human KLF factors are indicated. The cysteine and histidine residues involved in zinc coordination are highlighted in blue; residues contacting DNA are highlighted in yellow. The arrows point to residues making base-specific contacts thus recognizing KLF1-binding sites in the genome. Variants are color-coded: class 1, green; class 2, blue; class 3, red; class 4, black. See supplemental Table 1 for details and references.
Distinct classes of KLF1 variants defined by patient phenotypes
Several promoter variants have been described that may affect transcription of the KLF1 gene. However, most commonly, variants in KLF1 alter the protein-coding sequences (Figure 2 and supplemental Table 1, available on the Blood Web site). These variants can be divided into four functional classes: (1) variants with no or minor functional consequences, (2) hypomorphic variants with reduced function, (3) truncating loss-of-function variants, and (4) dominant variants. Class 1 represents missense variants located outside the DNA binding domain. These are regarded as neutral polymorphisms, although variants in the TADs might affect KLF1 activity. Class 2 comprises missense variants or small in-frame deletions that interfere with normal KLF1 function, which are almost invariably found within the DNA binding domain. Virtually all patients with compound heterozygous KLF1 variants carry a class 2 variant on at least one allele. Because the complete absence of functional KLF1 severely affects erythropoiesis,6,11,12 this illustrates that class 2 variants are hypomorphic alleles. Class 3 comprises stop codon or frameshift variants that result in truncated KLF1 proteins lacking the DNA-binding domain. Class 3 variants affecting only 1 allele cause haploinsufficiency for KLF1, which is phenotypically mild.1,2 Class 4 is represented by a single variant that changes a highly conserved residue in ZF2, p.E325K. It occurs exclusively as a de novo variant and causes a dominant severe congenital dyserythropoietic anemia (CDA IV; OMIM #613673).3,5
KLF1 variants underlie a wide range of red cell phenotypes
Blood groups
In(Lu) (inhibitor of Lutheran) is a clinically unremarkable blood group (OMIM #111150). The BCAM gene that encodes the Lutheran antigens was found to be intact in In(Lu) individuals, suggesting that BCAM expression was instead altered by variants in one of its regulatory proteins. In 2008, the first KLF1 variants were reported in 21 of 24 In(Lu) individuals.1 In all 21 cases, one normal KLF1 allele was present and the other allele carried a class 2 or 3 KLF1 variant. Expression of BCAM is highly sensitive to the level of functional KLF1; as a result, a reduced level of KLF1 presents as the In(Lu) phenotype.1 In combination with low CD44 (Indian blood group) expression, these markers provide useful flags for class 2 or 3 KLF1 variants.38 Many other blood group antigens are also direct KLF1 targets, but most are less sensitive to its levels.15,16,25,39,40 There is reduced expression of Kell, Duffy, Kidd, RhD, RhAG, Scianna, and LW blood group antigens in KLF1 null mice and in humans.6,14,15,18
Dysregulated globin expression
Since 1982, it has been known that variants in the promoter of the HBB gene (encoding β-globin) cause β+-thalassemia with elevated HbF.41,42 In 1994, these variants were shown to affect binding of KLF1,43 which suggested that KLF1 regulates the switch from fetal γ-globin to adult β-globin expression.44 Analysis of mice carrying a complete human HBB locus transgene and Klf1-null alleles supported this notion.45,46 In 2010, a family from Malta with hereditary persistence of HbF (HPFH) was reported in which the HPFH individuals carried a class 3 variant (p.K288X) on 1 allele of KLF1.2 Overexpression of full-length KLF1 in erythroid progenitors derived from such individuals corrected the phenotype, demonstrating that haploinsufficiency for KLF1 causes HPFH (OMIM #613566). Initially, it was thought that some individuals with class 2 or 3 KLF1 variants had In(Lu) whereas others had HPFH. We now know that carriers of class 2 or 3 KLF1 variants display both phenotypes. Compound heterozygotes for class 2 and 3 KLF1 variants display very high HbF levels of up to 40% of total hemoglobin (Hb),47 often accompanied by persistent expression of embryonic globins.4
In β-thalassemia carriers, the level of a minor Hb (HbA2; α2δ2) is increased and is diagnostic of carrier status.48 Class 2 or 3 KLF1 variants are associated with moderately increased levels of HbA2.7,8 Thus, as part of screening programs for thalassemia, sequencing of KLF1 is now strongly recommended in cases of borderline raised HbA2, especially if accompanied by raised HbF.
Iron and heme
KLF1 coordinates expression of many of the genes involved in iron metabolism of erythroid precursors, including heme synthesis enzymes (eg, ALAS2, ALAD, HMBS)14-16,49 and proteins regulating the processing of iron (eg, TFR2, SLC25A37, STEAP3, ABCG2, and ABCB10).6,19 In patients with class 2 or 3 KLF1 variants, the iron stores are usually normal and yet iron is not effectively incorporated into heme. In this situation, zinc rather than iron may be incorporated into heme. Thus, elevated ZnPP in the presence of normal iron stores is another useful flag for the presence of a class 2 or 3 KLF1 variant.47,50
Pyruvate kinase deficiency
KLF1 activates the PKLR gene encoding PK; PK levels are pathologically reduced in compound heterozygotes for class 2 and 3 KLF1 variants.4 These individuals display abnormal red blood cells reminiscent of prickle cells typical for PK deficiency. Red blood cell inclusions (siderocytes, Howell-Jolly bodies, and Pappenheimer bodies), schistocytes, and microcytes are also present in the blood films of some patients.50 In fact, there is no typical red cell appearance with KLF1 deficiency but rather a broad spectrum of morphologies that differ depending upon the underlying variants. Because KLF1 regulates many of the genes implicated in red cell enzyme deficiencies,4,14-16,19,49 routine enzyme assays can easily lead to misdiagnosis.
Nonspherocytic hemolytic anemia
Nonspherocytic hemolytic anemia (NSHA) is a label applied to inherited anemias characterized by shortened survival of red blood cells that have abnormal morphology (schistocytes but few spherocytes), erythroid hyperplasia in the marrow, and evidence of hemolysis (low haptoglobins, increased bilirubin, and increased lactate dehydrogenase). The label is usually applied once thalassemia, hereditary spherocytosis, enzymopathies, and CDA have been excluded by molecular, enzyme, and morphologic tests. Patients who are compound heterozygous for class 2 and 3 KLF1 variants often have a disease best labeled as NSHA.4,50 The class 2 p.A298P variant is commonly encountered in Asia; when inherited with class 3 KLF1 alleles, this variant leads to severe NSHA.4,50 Presumably, class 2 KLF1 variants have altered DNA-binding properties that affect gene expression in a variant-specific manner. Further work is required to fully address the mechanisms of action of individual class 2 variants. Potential mechanisms include selective loss of binding to specific target sequences in the genome, a generally reduced affinity that affects binding to all in vivo DNA-binding sites, reduced specificity that leads to off-target interactions, and altered protein-protein interactions with transcriptional coregulators.
CDA
CDA is a rare disorder of erythropoiesis that includes four subtypes. One missense variant (p.E325K) that changes a highly conserved residue in ZF2 of KLF1 (Figure 2) has a strong dominant negative effect and causes CDA IV (OMIM #613673).3,5,51 This variant has appeared de novo in at least 6 independent cases. The phenotype is much more severe than those caused by class 2 or 3 KLF1 variants and shows NSHA with marked erythroblastosis, binuclear erythroblasts in the marrow and circulation, and abnormalities typical of CDA that are visible with electron microscopy.3 In addition, CDA IV is characterized by very high expression of HbF (∼35% of total Hb) and persistent expression of embryonic globins.3 Notably, a variant in the homologous residue in mouse KLF1, p.E339D, underlies semidominant neonatal anemia (Nan).52,53 Phenotypically, Nan mice display many similarities with CDA IV patients, and a limited subset of KLF1 target genes is downregulated in Nan mice, which may be instructive for analysis of CDA IV patients.
Hydrops fetalis
From observations in mice, it could be predicted that homozygosity for class 3 KLF1 variants would invariably lead to fetal lethality. Remarkably, a human KLF1-null neonate was recently reported6 ; this child, whose parents were apparently normal, was born with severe anemia, jaundice, and fetal distress and was transfusion-dependent from birth. There was persistent HbF expression. Cerebral palsy occurred probably as a result of kernicterus. It transpired that both parents were carriers of class 3 KLF1 variants: frameshift variant p.R319Efs34X and stop-codon variant p.W30X. Transcriptome analysis showed that the pattern of altered gene expression in erythroid cells is very similar to that reported for Klf1-null mice.6,15,16 It is not clear whether the persistence of HbF explained survival to birth or whether this infant had complementary variants that ameliorated the effects of KLF1 deficiency. Given the prevalence of class 3 KLF1 variants in some populations,7,54 it is inevitable that other KLF1-null cases have escaped detection. To provide appropriate management at the earliest possible stage, it is important to detect these cases prenatally.
Prevalence of KLF1 variants and interactions with hemoglobinopathies
Until recently, KLF1 variants were considered to be extremely rare causes of red cell disorders. The identification of numerous sporadic cases by high throughput DNA sequencing prompted population surveys. In Southern China, the combined incidence of class 2 and 3 KLF1 variants (1.3%) is remarkably high, whereas in Northern China it is low, which correlates with the distribution of hemoglobinopathies in these regions. Consequently, co-inheritance of hemoglobinopathies and KLF1 variants is common7,55 and is likely to be the case in other areas where hemoglobinopathies are endemic such as the Mediterranean8,47 and Southeast Asia.4,55-57 The incidence of KLF1 mutations in Africa has not been tested, although there are reports of KLF1 variants in patients of African descent.58 Sequence analysis of KLF1 in 32 samples representing the lower and upper 10th percentiles of the HbF values from 250 patients with sickle cell diseases indicates that KLF1 variants are not commonly associated with increased HbF in populations of African descent (Swee-Lay Thein, unpublished data). Genetic traits that partially prevent switching from γ-globin to β-globin expression ameliorate the clinical severity of β-hemoglobinopathies.59 KLF1 positively regulates Hb switching,2,45 and switching is impaired in carriers of class 2 or 3 KLF1 variants such that higher HbF expression persists throughout life.54 Thus, in β-hemoglobinopathies, clinical benefit is derived from an additional class 2 or 3 KLF1 variant (Figure 3). Remarkably, expression of HbF remains very high (>30% of total Hb) in β-thalassemia patients with compound heterozygosity for class 2 and 3 KLF1 variants.47,50 Such levels of HbF significantly ameliorate the disease severity.
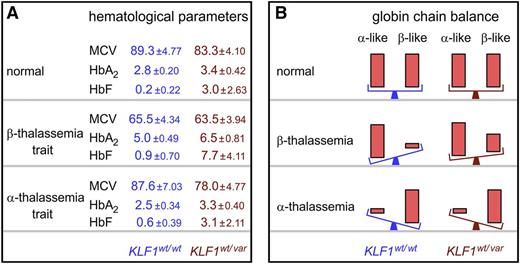
Impact of class 2 or 3 KLF1 variants on clinical severity of hemoglobinopathies. (A) Three major hematologic indices— mean cell volume (MCV), HbA2, and HbF—in controls and α- or β-thalassemia trait individuals with (wt/var) or without (wt/wt) class 2 or 3 KLF1 variants. Values are mean ± standard deviation, derived from Liu et al7 and Yu et al.55 (B) Effects of class 2 or 3 KLF1 variants on globin chain balance in healthy controls (normal) and α- or β-thalassemia patients. Significant recovery of globin chain balance is observed only when β-thalassemia is co-inherited with a class 2 or 3 KLF1 variant. α-like denotes ζ- and α-globin chains, and β-like denotes ε-, γ-, δ-, and β-globin chains. wt, wild type; var, variant.
Impact of class 2 or 3 KLF1 variants on clinical severity of hemoglobinopathies. (A) Three major hematologic indices— mean cell volume (MCV), HbA2, and HbF—in controls and α- or β-thalassemia trait individuals with (wt/var) or without (wt/wt) class 2 or 3 KLF1 variants. Values are mean ± standard deviation, derived from Liu et al7 and Yu et al.55 (B) Effects of class 2 or 3 KLF1 variants on globin chain balance in healthy controls (normal) and α- or β-thalassemia patients. Significant recovery of globin chain balance is observed only when β-thalassemia is co-inherited with a class 2 or 3 KLF1 variant. α-like denotes ζ- and α-globin chains, and β-like denotes ε-, γ-, δ-, and β-globin chains. wt, wild type; var, variant.
In α-thalassemia, there is no clinical benefit derived from an additional class 2 or 3 KLF1 variant.55 There is a mild but statistically significant decrease in mean cell volume in the presence of a class 2 or 3 KLF1 variant, but this is not noticeable in routine diagnostics (Figure 3).55 KLF1 directly activates the HBB gene,11,12,45 and partial correction of the α-globin:β-globin chain ratios would be expected to ameliorate disease severity. This has not been observed, presumably because of deregulation of other KLF1 target genes such as those required for iron metabolism21 and heme biosynthesis14,19,49 and the α-globin chaperone AHSP.14,17
Intriguingly, none of these effects is likely to provide an evolutionary advantage. Throughout human history, most individuals with severe hemoglobinopathies died before reaching reproductive age. Most KLF1 variants would have been neutral or detrimental, and even the variants that caused increased HbF would have been beneficial only when co-inherited with a β-hemoglobinopathy. So why did KLF1 variants reach polymorphic frequencies in some populations? Because class 2 or 3 KLF1 variants produce widespread changes in the structure and function of red blood cells, it seems likely that they create a suboptimal environment for the propagation of malaria parasites, thus providing a survival advantage. This is in keeping with the historical selection of hemoglobinopathies, red cell enzymopathies, and membrane defects in malaria-infested areas and with the geographic pattern of KLF1 variants that is now emerging.
Diagnosis of individuals with KLF1 variants
Carriers of class 2 or 3 KLF1 variants cannot be detected via routine full blood examinations. Mean cell volume, and mean corpuscular hemoglobin are statistically lower than in controls, but the indices of most carriers fall within the low end of the normal range.7 To complicate matters further, there are many KLF1 gene variants of unknown significance that should nevertheless be reported in the public domain (eg, the HbVar database for red cell disorders at http://globin.bx.psu.edu/hbvar60 ) with microattribution credits to the authors.61 An integrated set of comprehensive locus-specific databases for gene variants involved in red cell disorders, including KLF1, can help identify genomic variants with functional significance, whereas the identification of common neutral variants may rule out a role in disease. Similarly, genomic variants of key regulatory regions could be critical in identifying important cis-elements and mutational mechanisms such as gene conversion events. Conversely, neutral variants may help identify regions of little or no functional significance. Thus, microattribution may pose and answer questions that would otherwise not be addressed, potentially leading to useful new insights while providing credit to new data submitters. Recently acquired knowledge on genotype-phenotype associations of class 2 or 3 KLF1 variants suggests that several additional tests could be performed to evaluate the likelihood of an individual harboring such a variant. Increase in reticulocytes and HbF, borderline increase in HbA2, increased ZnPP with normal iron stores, reduced CD44 surface expression, and the In(Lu) blood group are all flags for the presence of a class 2 or 3 KLF1 variant (Figure 4). Sequencing the three KLF1 exons and the promoter is straightforward and appropriate if carrier status is suspected. On the basis of previous studies, it is certain that many carriers of class 2 or 3 KLF1 variants remain undetected in at-risk populations. The chance of a KLF1-null child being conceived is significant (eg, 1 in every 24 000 conceptions in Southern China).7 Although most KLF1-null fetuses probably would die in utero, some may be born alive with severe hydrops fetalis.6 Unlike with the common hemoglobinopathies, this condition will not be anticipated unless an affected family member has previously been identified. Some of us have recently undertaken chorionic villus sampling and KLF1 sequencing for prenatal diagnosis in an affected family. The parents carried class 3 variants and already had a severely affected KLF1-null child, so the counseling was relatively straightforward. Careful genetic counseling is required when compound homozygosity for class 2 and 3 KLF1 variants is detected because the child’s phenotype will depend on the remaining functionality of the class 2 KLF1 variant.
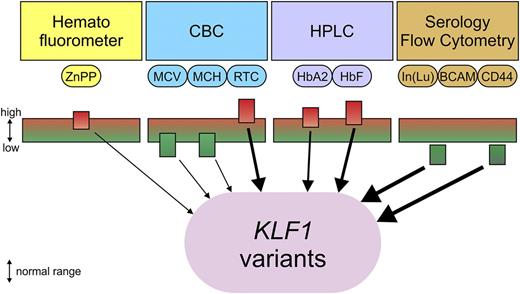
Flags indicating carrier status of class 2 or 3 KLF1 variants. Automated complete blood count (CBC): mean cell volume (MCV) and mean corpuscular hemoglobin (MCH) are low or at the low end of the normal range. Reticulocyte count (RTC) is high or at the high end of the normal range. ZnPP measured by hematofluorometer is high or at the high end of the normal range, with normal iron stores and absence of lead poisoning. HbA2 and HbF are measured by high-performance liquid chromatography (HPLC) and are moderately increased or at the high end of the normal range. Lutheran antigens are measured by serology and are decreased but not completely absent; Indian antigen (CD44) is measured by flow cytometry and is reduced with normal expression of glycophorin A (CD235a). Weight of the arrows indicates the likelihood that a class 2 or 3 KLF1 variant is present; this increases when several flags are observed simultaneously.
Flags indicating carrier status of class 2 or 3 KLF1 variants. Automated complete blood count (CBC): mean cell volume (MCV) and mean corpuscular hemoglobin (MCH) are low or at the low end of the normal range. Reticulocyte count (RTC) is high or at the high end of the normal range. ZnPP measured by hematofluorometer is high or at the high end of the normal range, with normal iron stores and absence of lead poisoning. HbA2 and HbF are measured by high-performance liquid chromatography (HPLC) and are moderately increased or at the high end of the normal range. Lutheran antigens are measured by serology and are decreased but not completely absent; Indian antigen (CD44) is measured by flow cytometry and is reduced with normal expression of glycophorin A (CD235a). Weight of the arrows indicates the likelihood that a class 2 or 3 KLF1 variant is present; this increases when several flags are observed simultaneously.
Wider implications and future directions
Here we have illustrated how high-throughput DNA sequencing can directly inform clinical practice. This approach has followed on from a variety of laborious approaches used to initially diagnose rare cases of KLF1 variants. By using DNA sequencing as the primary analytical tool, we have now uncovered an important disease gene that unexpectedly accounts for a significant proportion of unexplained red blood cell disorders. This critical new knowledge has raised the dilemma of whether to and how to screen for such variants in populations at risk.
Why was this important disease gene not discovered earlier? First, carriers are not readily detected by routine diagnostic tests. Second, with focused genetic studies, KLF1 variants were not considered as a possible cause of phenotypes such as PK deficiency. Until high-throughput DNA sequencing became widely used, such cases of unexplained anemia remained in abeyance. Third, variants in master regulators cause a wide range of loosely associated phenotypes that will differ depending on the specific variants present in the patient. We note that Klf1 mouse models have been highly informative for understanding the human phenotypes.23,62,63 Recent advances in genome editing technology have enabled the introduction of patient-specific variants in mice at an unprecedented scale.64,65 Such mice will be invaluable tools for investigating the impact of these variants at the physiologic, cellular, and molecular level. This detailed knowledge is essential for providing the best possible counseling and clinical care.
We predict that variants affecting other master regulators of key cell types will account for conditions with hitherto unexplained genetics. These currently enigmatic diseases most likely share features of several conditions affecting the particular organ system. As in KLF1, variants in some of these genes may turn out to be much more frequent than anticipated. The ever-reducing threshold for whole genome sequencing promises to reveal new examples in a wide range of organ systems in the near future.
The online version of this article contains a data supplement.
Acknowledgments
This work was supported by the Landsteiner Foundation for Blood Transfusion Research (LSBR 1040), The Netherlands Organization for Scientific Research (NWO/ZonMw TOP 40-00812-98-12128), and the EU Seventh Framework Program Specific Cooperation Research Project THALAMOSS (306201) (S.P.); EU FP7 HEALTH projects (200754 and 305444) (G.P.P.); Medical Research Council UK and the National Institute for Health Research Oxford Biomedical Research Centre Programme (D.R.H.); National Natural Science Foundation of China-Guangdong Joint Fund (No. U1201222) and National Key Technology Research and Development Program China (No. 2012BAI09B01) (X.X.); National Health Medical Research Council APP1082439 Australia (A.P.); National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases grants R01-DK046865 and R01-DK102260, and New York Stem Cell Foundation grant CO26435 (J.J.B.).
Authorship
Contribution: A.P., X.X., D.R.H., G.P.P., L.A., J.J.B., and S.P. coordinated writing of the review; A.P., X.X., and S.P. prepared the figures; S.P. prepared supplemental Table 1; and all authors contributed to the article by revising initial draft versions.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
A list of KLF1 Consensus Workgroup members can be found in the supplemental Data.
Correspondence: Sjaak Philipsen, Erasmus University Medical Center Rotterdam, Department of Cell Biology, Room Ee1000, PO Box 2040, 3000 CA Rotterdam, The Netherlands; e-mail: j.philipsen@erasmusmc.nl.