Key Points
CD45-driven expression of Cre generates the first mouse model that allows specific and exclusive deletion of Pten in hematopoietic cells.
Pten deletion in CD45-expressing cells causes T-cell acute lymphoblastic leukemia, but no other hematologic malignancies.
Abstract
Since its discovery in the late 1990s, Pten has turned out to be one of the most important tumor suppressor genes. Pten loss results in increased activation of the phosphatidylinositol 3-kinase/Akt signaling pathway, which is associated with increased proliferation, survival, and neoplastic growth. Here, we have addressed the effects of conditional deletion of Pten in hematopoietic cells by crossing Pten conditional knockout mice with a knock-in mouse expressing the Cre recombinase in the CD45 locus. CD45 is also known as leukocyte common antigen, and it is expressed in virtually all white cells and in hematopoietic stem cells. Using a reporter mouse, we demonstrate that CD45:Cre mouse displays recombinase activity in both myeloid and lymphoid cells. However, deletion of Pten in CD45-expressing cells induces development of T-cell acute lymphoblastic leukemia and lymphoma, but not other hematologic malignancies.
Introduction
Pten (phosphatase and tensin homolog deleted on chromosome 10) is a tumor suppressor gene that antagonizes the activation of the phosphatidylinositol 3-kinase/Akt signaling pathway. Either Pten loss or inactivation causes increased cell growth, proliferation, and survival.1 Pten can be altered in up to 80% of patients, depending on the type of cancer.2 Loss-of-function mutations of Pten are frequently found in solid tumors such as glioblastomas; thyroid, prostate, or endometrial carcinomas; and in hematologic malignancies.3
The role of Pten in maintenance of hematologic homeostasis has been evidenced by Pten knockout mouse models. Mice hemizygous for Pten (Pten+/−) develop multiple neoplasias.4-6 Development of mouse models that allow Pten loss in hematopoietic stem cells (HSCs) has been a breakthrough in the field.7-12 Collectively, the use of conditional Pten mouse models has revealed that Pten deletion leads to development of both acute myeloid leukemia and T-cell acute lymphoblastic leukemia/lymphoma (T-ALL).
To date, there are no existing mouse models to achieve specific ablation of Pten in hematopoietic cells without affecting other cell types. Here, we have assessed the effects of conditional deletion of Pten in CD45-expressing cells. Specific Pten loss in CD45-expressing cells led to development of T-ALL, but no other types of hematologic malignancies. Our results present a new mouse model for the study of Pten-induced hematologic malignancies.
Study design
Mice were housed and maintained as previously described.13 CD45:Cre mice were kindly donated by Dr Alexander Medvinsky. Breeding schemes are depicted in supplemental Figure 1 (available on the Blood Web site).
Western blot, histopathology and immunohistochemical analysis, and assessment of cell proliferation were performed as previously described.13,14 Antibodies and dilutions are shown in the supplemental Methods.
Cytological analysis was performed with blood collected from the submandibular vein. Smears were immediately prepared and fixed in 100% ethanol. For Papanicolau staining, ethanol-fixed slides were stained with hematoxylin and counterstained with Orange G and Eosin Azure (EA50), cleaned with 95% ethanol, and mounted.
Flow cytometry was performed on bone marrow, spleen, and blood cells. Fluorescence emission was measured using BD FACS Canto II (BD Biosciences, San Jose, CA), and population distribution was analyzed with FACSDiva Software (BD Biosciences). For detailed protocol and antibodies used, see supplemental Methods.
Recombinase activity reporter assays were performed by crossing CD45:Cre+/− and mT/mGfl/+ mice (supplemental Figure 1). The resulting offspring was analyzed as previously described.13
Statistical analysis
All the experiments were performed and repeated at least 3 times. N indicates the number of mice. Statistical analyses were performed using Prism 6.0 software (GraphPad). Statistical significance was evaluated using analysis of variance analysis followed by Bonferroni’s test for multiple comparisons. P ≤ .05 was considered statistically significant.
Results and discussion
CD45 encodes a receptor type protein designated as the leukocyte common antigen that is expressed in all nucleated hematopoietic cells, including HSCs and mature lymphoid and myeloid cells. In order to test the recombinase activity, we crossed CD45:Cre mice15 with double fluorescent reporter mice (mT/mG).16 Five weeks after birth, the offspring CD45:Cre+/− mT/mGf/− was analyzed for the presence of red and green fluorescence in different hematopoietic organs (supplemental Figure 2A). We observed the presence of green leukocytes in all the tissues, suggesting efficient recombination in both myeloid and lymphoid subsets. To confirm these results, we analyzed the percentage of monocytes (Mac-1+ Gr1−), granulocytes (Mac-1+ Gr1+), T cells (CD3+), and B lymphocytes (B220+) displaying green fluorescence in peripheral blood (supplemental Figure 2B-C). GFP+ cells represented >75% of monocytes and B and T cells, whereas in granulocytes recombination occurred in ∼40% of the population (supplemental Figure 2D). These results evidenced that the expression and activity of CD45:Cre was able to cause recombination in all mature blood lineages, although different efficiencies were observed depending on the population.
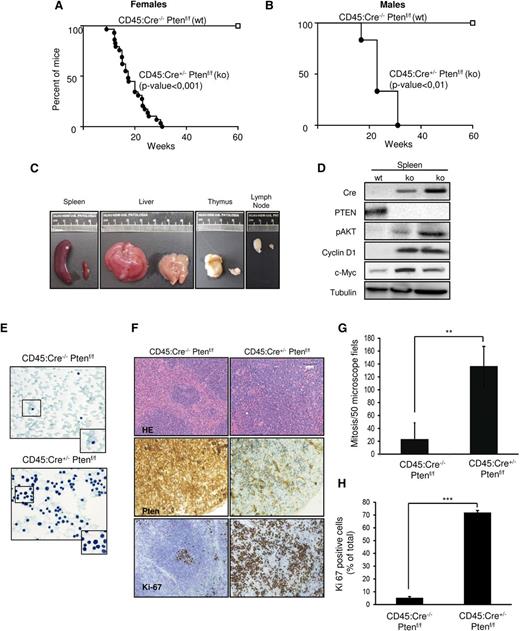
Although different mouse models have been developed to study the role of Pten in leukemogenesis, all of them lead to Pten deletion in other cell types besides leukocytes. Here, we describe the first genetic approach to achieve Pten ablation exclusively in hematopoietic tissue. To interrogate the impact of Pten loss in the hematopoietic compartment, conditional Pten knockout mice (Ptenf/f)17 were bred with CD45:Cre+/−15 (supplemental Figure 1). CD45:Cre+/− Ptenf/f animals displayed a dramatically reduced life span (Figure 1A-B). Necropsy of mice revealed presence of splenomegaly, hepatomegaly, severe thymus enlargement, and lymphadenopathy in all Pten-deficient animals (Figure 1C), suggesting development of lymphomas. Moreover, recombinase expression and Pten loss was confirmed in spleens by western blot analysis (Figure 1D), correlating with an increase in Akt phosphorylation, as well as an increment of cyclin D1 and c-Myc expression, both of them targets of Pten in lymphomagenesis.
CD45:Cre+/−Ptenf/f develop lethal disease that affects lymphoid organs. (A-B) Kaplan-Meier curve of females and males analyzed for 60 weeks. Note that survival dramatically decreases after 20 weeks. (C) Macroscopic comparison of representative spleen, liver, thymus, and lymph nodes from CD45:Cre+/−Ptenf/f (ko, left) and CD45:Cre−/−Ptenf/f (wt, right). (D) Western blot from wt and ko spleens showing Cre recombinase expression, correlating with Pten loss and increased Akt phosphorylation (p-Akt), cyclin D1, and c-Myc levels. Membranes were reprobed with tubulin to ensure protein loading. (E) Representative images (×63) showing Papanicolau staining of blood extensions from CD45:Cre−/−Ptenf/f and CD45:Cre+/−Ptenf/f mice. (F) Representative images (×10) showing hematoxylin and eosin (H&E) staining and Pten and Ki-67 immunohistochemistry of spleens from CD45:Cre−/−Ptenf/f and CD45:Cre+/−Ptenf/f mice. (G-H) Quantification of mitotic activity and proliferation in spleens from CD45:Cre−/−Ptenf/f and CD45:Cre+/−Ptenf/f mice. Results are expressed as number of mitotic figures observed per 50 high-power microscope fields (G) and percent of total cells displaying nuclear staining for Ki-67 (H). Data are means ± standard deviation; **P ≤ .001; ***P ≤ .0001.
CD45:Cre+/−Ptenf/f develop lethal disease that affects lymphoid organs. (A-B) Kaplan-Meier curve of females and males analyzed for 60 weeks. Note that survival dramatically decreases after 20 weeks. (C) Macroscopic comparison of representative spleen, liver, thymus, and lymph nodes from CD45:Cre+/−Ptenf/f (ko, left) and CD45:Cre−/−Ptenf/f (wt, right). (D) Western blot from wt and ko spleens showing Cre recombinase expression, correlating with Pten loss and increased Akt phosphorylation (p-Akt), cyclin D1, and c-Myc levels. Membranes were reprobed with tubulin to ensure protein loading. (E) Representative images (×63) showing Papanicolau staining of blood extensions from CD45:Cre−/−Ptenf/f and CD45:Cre+/−Ptenf/f mice. (F) Representative images (×10) showing hematoxylin and eosin (H&E) staining and Pten and Ki-67 immunohistochemistry of spleens from CD45:Cre−/−Ptenf/f and CD45:Cre+/−Ptenf/f mice. (G-H) Quantification of mitotic activity and proliferation in spleens from CD45:Cre−/−Ptenf/f and CD45:Cre+/−Ptenf/f mice. Results are expressed as number of mitotic figures observed per 50 high-power microscope fields (G) and percent of total cells displaying nuclear staining for Ki-67 (H). Data are means ± standard deviation; **P ≤ .001; ***P ≤ .0001.
Histopathological examination of blood extensions revealed the presence of enlarged lymphoid cells that exhibited increased and irregular nuclei and the presence of high number of mitotic cells (Figure 1E). Histopathological analysis of the spleen revealed disruption of its normal architecture by a diffuse infiltration of lymphoid cells (Figure 1F) that exhibited irregular nuclei, prominent nucleoli, and high mitotic rates (Figure 1G), as well as cytological atypia and strong Ki-67 immunoreactivity (Figure 1H). Histologic evaluation of lungs, liver, colon, and kidneys confirmed infiltration of atypical lymphoid cells lacking Pten (supplemental Figure 3). All these observations indicate that Pten loss in CD45-expressing cells results in development of severe lymphomas/leukemia.
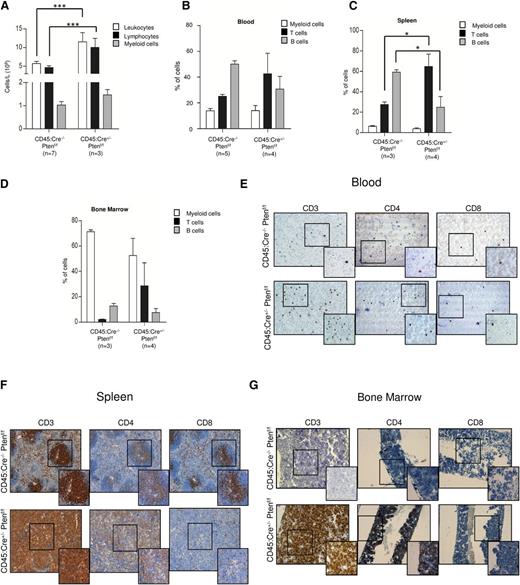
Next, we investigated whether Pten loss was causing myeloid or lymphoid disease in CD45:Cre+/− Ptenf/f mice. Complete blood cell count from 6-month-old sick animals revealed a significant increase in the number of leukocytes, because of an increase in lymphocytes (Figure 2A). Flow cytometry analysis showed a marked increment in the percentage of T cells accompanied by a reduction of B lymphocytes in peripheral blood, spleen, and bone marrow from 4-month-old mice (Figure 2B-D) indicating that the developed malignancies were compatible with T-ALL. Strikingly, none of the mice analyzed displayed features of myeloid or B-cell malignancies. The T-cell nature of the disease was further proved by immunohistochemical analysis of blood, bone marrow, and spleen. In CD45:Cre+/− Ptenf/f animals, blood, bone marrow, and the germinal centers of spleen were invaded by CD3+ CD4+ CD8− T cells (Figure 2E-G). Consistently, Pten-negative lymphocytic infiltrates found in lung, liver, colon, and kidney were positive for CD3 (supplemental Figure 3). In summary, these data demonstrate that Pten loss in CD45+ cells leads to development of T-cell lymphoma/leukemia but not to other malignancies.
Pten loss in CD45-expressing cells leads to development of T-ALL. (A) Complete blood cells counts showing total number of leukocytes, lymphocytes, and myeloid cells in CD45:Cre−/−Ptenf/f and CD45:Cre+/−Ptenf/f mice. (B-D) Quantification of flow cytometry analysis of blood (B), spleen (C), and bone marrow (D) from CD45:Cre−/−Ptenf/f and CD45:Cre+/−Ptenf/f mice. (E-G) Representative images of CD3, CD4, and CD8 immunhistochemistry in blood (E), spleen (F), and bone marrow (G) from CD45:Cre−/−Ptenf/f and CD45:Cre+/−Ptenf/f mice. Data are means ± standard deviation; *P ≤ .05; ***P ≤ .0001.
Pten loss in CD45-expressing cells leads to development of T-ALL. (A) Complete blood cells counts showing total number of leukocytes, lymphocytes, and myeloid cells in CD45:Cre−/−Ptenf/f and CD45:Cre+/−Ptenf/f mice. (B-D) Quantification of flow cytometry analysis of blood (B), spleen (C), and bone marrow (D) from CD45:Cre−/−Ptenf/f and CD45:Cre+/−Ptenf/f mice. (E-G) Representative images of CD3, CD4, and CD8 immunhistochemistry in blood (E), spleen (F), and bone marrow (G) from CD45:Cre−/−Ptenf/f and CD45:Cre+/−Ptenf/f mice. Data are means ± standard deviation; *P ≤ .05; ***P ≤ .0001.
Previous studies have demonstrated that Pten is required to maintain the pool of HSCs, playing a role in lineage choice and leukemogenesis. Pten deletion using Mx1-Cre mice leads to a rapid formation of myeloproliferative disease that mainly progresses to acute myeloid leukemia when Pten is lost in adult mice 8 weeks after birth.12 When the deletion happens at 3 weeks of age, mice die of myeloproliferative disease in adulthood.18 In contrast, mouse models including VE-cadherin-Cre,8 Vav-iCre,9 and Scl-CreERT,10 where Pten is lost during fetal development or early after birth, show that Pten loss leads to T-ALL but no other malignancies. Our model is closer to the second group, as CD45 is expressed already in the fetal stage of hematopoiesis. All together, these facts suggest that the phenotype of the pathology depends not only on the gene affected, but also on the moment when the mutation occurs. When Pten loss occurs during development it leads to T-ALL, but when it takes place in adulthood it induces myleoid malignancies. It is noteworthy that none of the previous models delete Pten exclusively in hematopoietic cells. Thus, deletion of Pten in other cell types could affect the behavior of hematopoietic cells. We report the first study in which Pten has been knocked out in hematopoietic cells since early development, without affecting other cell types and avoiding the use of substances that can modify hematopoiesis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Mónica Domingo and Montse Ortrega for their technical support.
This work was supported by grants from Fondo de Investigaciones Sanitarias del Instituto de Salud Carlos III cofinanciado por Fondo Europeo de Desarrollo Regional (“Una manera de hacer Europa”) (PI13/00263 and PI13/01701), Red Temática de investigación en Cáncer (RD06/0020/1034), Grups consolidats de la Generatitat de Catalunys (2009SGR794), Fundació La Marato de TV3, Grupos estables Asociación Española Contra el Cancer, Catalunya contra el cáncer and programa de intensificación de la investigación, Instituto Carlos III.
The funders had no role in study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
Authorship
Contribution: C.M., M.A.D., X.M.-G., and X.D. conceived and designed the experiments; C.M., N.E., and M.A.D. performed the experiments; D.H., M.S., and J.Y. provided technical support; S.G., F.V., and X.M.-G. analyzed the data; and C.M., A.M., and X.D. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Xavi Dolcet, Departament de Ciències Mèdiques Bàsiques, Universitat de Lleida/IRBLleida, Ed Biomedicina I, Hospital Arnau de Vilanova, Ave Rovira Roure 80, 25198 Lleida, Spain; e-mail: dolcet@cmb.udl.cat.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal