In this issue of Blood, Scarfò et al describe a novel subset of anaplastic lymphoma kinase (ALK)-negative anaplastic large-cell lymphoma (ALCL) associated with aberrant expression of ERBB4 transcripts and potential clinical relevance.1
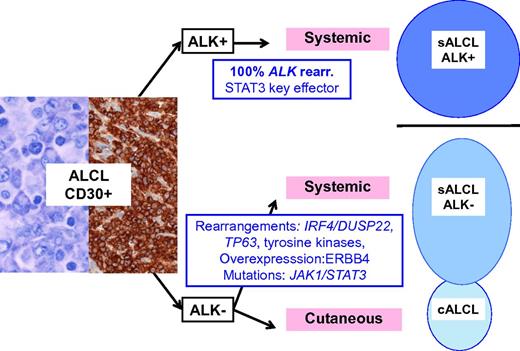
Schematic representation of ALCL entities and their genetic-associated aberrations. ALK-positive ALCL is a well-delineated entity, primarily defined by ALK translocations. Other lymphomas with similar anaplastic morphology and CD30 expression do occur, either as a systemic disease, referred as ALK-negative ALCL, a provisional entity in the current classification, or as a primary cutaneous neoplasm, primary cutaneous ALCL, which are part of the spectrum of primary cutaneous CD30+ lymphoproliferative disorders and usually pursue a benign course. sALCL, systemic ALCL; rear., rearrangement.
Schematic representation of ALCL entities and their genetic-associated aberrations. ALK-positive ALCL is a well-delineated entity, primarily defined by ALK translocations. Other lymphomas with similar anaplastic morphology and CD30 expression do occur, either as a systemic disease, referred as ALK-negative ALCL, a provisional entity in the current classification, or as a primary cutaneous neoplasm, primary cutaneous ALCL, which are part of the spectrum of primary cutaneous CD30+ lymphoproliferative disorders and usually pursue a benign course. sALCL, systemic ALCL; rear., rearrangement.
ALCLs represent a subset of peripheral T-cell lymphomas (PTCLs) defined by a proliferation of large lymphoid cells, referred to as hallmark cells, with strong expression of CD30. The molecular deciphering of ALCL started in the 1990s, with the discovery of a recurrent t(2;5)(p23;q35) translocation fusing the ALK gene and the nucleophosmin (NPM) gene generating a NPM-ALK fusion protein in a subset of ALCL, and subsequent description of alternative ALK translocations resulting in high ALK kinase activity.2 NPM-ALK triggers proliferation and survival pathways and represents the major oncogenic driver in ALK-positive ALCL. Accordingly, pharmacologic ALK inhibition has shown efficacy in relapsed/refractory ALK-positive ALCL patients.3 Compared with ALK-negative cases, ALK-positive ALCL occurs in younger patients and has a better prognosis.2,3 Consequently, in the last 2008 World Health Organization (WHO) classification, ALK-positive ALCL has been individualized as a distinct disease entity whereas ALK-negative ALCL, defined by a morphology and phenotype similar to ALK-positive ALCL but lacking ALK rearrangement and ALK expression, is listed as a provisional PTCL entity.4 Although ALK-negative ALCL is clearly nonoverlapping with ALK-positive ALCL, conversely, its distinction from other PTCL-expressing CD30 may be more subjective. Importantly, this question is of clinical relevance because the prognosis of ALK-negative ALCL and CD30+ PTCL–not otherwise specified (NOS) differs, but it still remains a matter of debate.3
Although gene expression profiling studies have shown that ALK-positive and ALK-negative ALCL share a common signature, suggesting a common cellular origin and/or shared pathogenic pathways,5 the driver genetic alterations in ALK-negative ALCL remained until recently poorly characterized. With the development of next-generation sequencing technologies, an increasing number of genetic aberrations have emerged in ALK-negative ALCL. Rearrangements at 6p25.3 involving DUSP22, a gene encoding a dual-specificity phosphatase that inhibits T-cell receptor signaling, are reported in about 30% of the cases and result in DUSP22 downregulation.6 Interestingly, these rearrangements are found in systemic ALK-negative ALCL but also in primary cutaneous ALCL (cALCL) which constitutes a separate disease with distinct clinical features and an indolent outcome.7 TP63 rearrangements creating fusion proteins homologous to a dominant-negative p63 isoform define another discrete genetic subset (8% of the cases).6 In one study, the prognosis of DUSP22-rearranged and ALK-positive ALCLs was very similar whereas TP63-rearranged cases were associated with a poor outcome, suggesting that molecular subclassification may be clinically relevant.8 Molecular heterogeneity of ALK-negative ALCL was more recently emphasized by the demonstration of recurrent activating signal transducer and activator of transcription 3 (STAT3) or Janus kinase 1 (JAK1) mutations in around 20% of cases, and the presence of fusion transcripts involving tyrosine kinases (ROS1 or TYK2) in other cases.9 Interestingly, the latter aberrations lead to the constitutive activation of the JAK/STAT3 pathway, which is also a central feature of ALK-positive ALCL as a consequence of ALK signaling (see figure).
In this issue, using an effective algorithm designated cancer outlier profile analysis (COPA) applied to a gene expression data set including various PTCLs and normal T cells, Scarfò et al report on the identification of a novel subset of ALK-negative ALCL coexpressing ERBB4 and COL29A1 and featuring a specific gene signature.1 The authors focused on ERBB4, the fourth member of the tyrosine kinase receptor ERBB family, which includes EGFR (ERBB1) and HER2 (ERBB2), known to be deregulated and/or mutated in several cancer types. HER2 is amplified in breast cancers, defining a subset with an aggressive behavior, and EGFR mutations are detected in many cancers, especially in lung adenocarcinomas. Interestingly, these 2 alterations can be effectively targeted by specific therapies which have dramatically improved the patient outcome.10 However, this family of tyrosine kinase receptors was not previously reported to be involved in lymphomagenesis.
Here, the authors found ERBB4 expression in 24% of ALK-negative ALCL, but not in PTCL-NOS nor in ALK-positive ALCL. Interestingly, this ectopic expression resulted from 2 different truncated transcripts I20ΔERBB4 and I12ΔERBB4. ERBB4 protein is expressed in a phosphorylated and active form, and associated with MMP9 expression. This ERBB4 activity is not due to a genomic alteration, but is related to the promotion of the intronic transcription start site by derepression of long terminal repeats from endogenous retrovirus. It seems that I12ΔERBB4, present at lower level, shows the highest oncogenic potential. Finally, using in vitro and in vivo experiments, in particular an ERBB4-positive patient-derived xenograft model, the authors show that treatment with neratinib, a pan-HER inhibitor, partially impairs tumor growth (see figure).
These novel findings still raise several questions regarding these ERBB4-positive ALCLs. It is suggested that ERBB4 ALCL may constitute a distinct subclass among ALK-negative ALCLs. Interestingly, ERBB4-positive cases frequently displayed an unusual Hodgkin-like morphology, but ERBB4-positive ALCL patients did not differ from other ALK-negative ALCL cases in terms of survival. Further studies are needed to determine whether ERBB4-expressing ALCLs overlap with other genetic subsets of ALK-negative ALCL and are also present in primary cutaneous ALCL. Whether ERBB4-positive ALCL patients may benefit from specific therapies remains to be explored. Indeed, the only partial effect of the kinase inhibition on the tumor growth in the preclinical model may indicate the need for combination therapies in relapsed or refractory ERBB4-positive ALCL patients. The identification of such patients in the clinical practice is another issue: in the absence of reliable ERBB4 antibodies applicable for immunohistochemistry in the clinical arenas, molecular tests would be needed unless the value of MMP9 expression as an alternative biomarker is further confirmed. Finally, the mechanisms leading to ERBB4 aberrant expression, especially whether epigenetic deregulation is involved, need to be clarified and better understood. This is of particular interest because various mutations affecting epigenetic modifiers have been recently described in PTCLs.
The article by Scarfò et al highlights how novel bioinformatics algorithms applied to a gene expression data set help identify novel molecular subsets within apparently homogeneous diseases. The recognition of this subclass of ERBB4 expressing ALK-negative ALCL, potentially targetable, is a new step toward a better understanding of ALCL pathogenesis. These findings add to the molecular landscape of ALK-negative ALCL which appears to include multiple subgroups driven by different genetic aberrations.
Conflict-of-interest disclosure: The authors declare no competing financial interests.