Key Points
The intestinal microbiota regulates the steady-state survival and turnover kinetics of circulating neutrophils and inflammatory monocytes.
The cytoplasmic peptido-glycan sensor Nod1 relays microbial signals into IL-17A–dependent stimulation of myeloid cell persistence.
Abstract
Maintenance of myeloid cell homeostasis requires continuous turnover of phagocytes from the bloodstream, yet whether environmental signals influence phagocyte longevity in the absence of inflammation remains unknown. Here, we show that the gut microbiota regulates the steady-state cellular lifespan of neutrophils and inflammatory monocytes, the 2 most abundant circulating myeloid cells and key contributors to inflammatory responses. Treatment of mice with broad-spectrum antibiotics, or with the gut-restricted aminoglycoside neomycin alone, accelerated phagocyte turnover and increased the rates of their spontaneous apoptosis. Metagenomic analyses revealed that neomycin altered the abundance of intestinal bacteria bearing γ-d-glutamyl-meso-diaminopimelic acid, a ligand for the intracellular peptidoglycan sensor Nod1. Accordingly, signaling through Nod1 was both necessary and sufficient to mediate the stimulatory influence of the flora on myeloid cell longevity. Stimulation of Nod1 signaling increased the frequency of lymphocytes in the murine intestine producing the proinflammatory cytokine interleukin 17A (IL-17A), and liberation of IL-17A was required for transmission of Nod1-dependent signals to circulating phagocytes. Together, these results define a mechanism through which intestinal microbes govern a central component of myeloid homeostasis and suggest perturbations of commensal communities can influence steady-state regulation of cell fate.
Introduction
Each day, ∼1011 mature neutrophils emerge from the bone marrow and enter systemic circulation, comprising the most abundant leukocyte subset in the human bloodstream.1 These terminally differentiated phagocytes migrate swiftly into inflamed tissues following infection or injury, where they eliminate invading pathogens, release an array of inflammatory mediators, and help orchestrate subsequent phases of immunity.2,3 To maintain tissue and immune homeostasis in the absence of inflammation, steady-state production of neutrophils must be counterpoised by continuous elimination in the bone marrow, spleen, and liver.4,5 The uniquely brief lifespan of these cells, averaging <24 hours, is essential for replenishing the pool of circulating phagocytes and averting the vascular and tissue damage risked by their potent cytotoxic capacity.6,7 However, pathologically enhanced neutrophil turnover has been linked to congenital neutropenia and a wide range of immunodeficiency syndromes in humans, illustrating the stringent regulation of cell survival required to maintain immunologic fitness.8 Despite the central role played by cell turnover in regulating phagocyte homeostasis, the factors influencing neutrophil lifespan during health remain incompletely understood.
Steady-state turnover of neutrophils follows a program of spontaneous cell senescence, exit from circulation, and programmed cell death.9 Neutrophils that have resided longest in the bloodstream shed the surface selectin CD62L and enhance display of the chemokine receptor CXCR4, changes that precede extravasion and correspond with enhanced inflammatory capacity (termed neutrophil “aging”).10,11 Simultaneously, accumulating oxidative damage and metabolic dysregulation cooperate to trigger spontaneous apoptosis among neutrophils upon their exit from the bloodstream.12,13 Tight coordination of extravasion and cell death allows for efficient and immunologically quiescent clearance of expiring phagocytes. Although the mechanisms of cell death in neutrophils have long been subject to intensive study, previous work has focused largely on the diverse cell-intrinsic pathways governing constitutive neutrophil apoptosis.14,15 Comparatively less attention has been paid to extrinsic factors influencing neutrophil viability in the steady state.
Extension of neutrophil lifespan during inflammation highlights the ability of environmental signals to modulate phagocyte turnover kinetics. Numerous viral and bacterial pathogens prolong human and murine neutrophil survival locally to stabilize a replicative niche or to promote tissue damage that facilitates microbial growth.16-18 Moreover, enhanced viability can augment neutrophil responses to some bacterial infections, driven by proinflammatory cytokines like interleukin (IL) 1β, tumor necrosis factor α, and IL-17A.19,20 Neutrophils obtained from patients with systemic inflammatory diseases, including rheumatoid arthritis, antineutrophil cytoplasmic antibody (ANCA)–associated vasculitis, gout, and cystic fibrosis also exhibit delayed apoptosis.7 The stimulatory impact of microbial and cytokine signals are thought to expand the window in which recruited neutrophils can exert inflammatory function, strengthening their contribution to host defense or tissue damage. Whether similar signaling influences neutrophil lifespan in the absence of inflammation remains unclear.
A growing body of evidence suggests that signals emanating from the gut microbiota prime systemic innate immunity in health and regulate phagocyte homeostasis.21 Germ-free mice show steady-state reductions in the abundance of both neutrophils and mononuclear phagocytes, changes ascribed previously to defects in hematopoietic development.22-24 However, mature phagocytes from animals raised germ-free or treated with antibiotics also exhibit a range of functional defects, including impaired trafficking to tissues, production of reactive oxygen species, and bactericidal capacity.25-27 Given that the commensal flora can influence phagocyte function in the periphery, we hypothesized that steady-state microbial signals may enhance the lifespan of circulating neutrophils and that this effect may extend to other short-lived, circulating phagocytes.
Here, we used in vivo cell-tracking and ex vivo cell viability assays to demonstrate that a distinct, neomycin-sensitive cohort of intestinal microbiota continuously regulates the death and turnover rates of neutrophils and Ly6C+ inflammatory monocytes (IMs) in healthy mice. We show that signaling through nucleotide-binding oligomerization domain-containing protein 1 (Nod1), a cytoplasmic peptidoglycan sensor that recognizes the dipeptide structure γ-d-glutamyl-meso-diaminopimelic acid (iE-DAP), is necessary and sufficient to mediate the microbial influence on phagocyte longevity. We find that stimulation of Nod1 signaling expands the number of lymphocytes producing the inflammatory cytokine IL-17A at the murine intestine. Finally, we demonstrate that IL-17A is required to transmit the Nod1-dependent microbial signal to neutrophils and IMs throughout the body, illuminating a signaling axis through which commensal microbes regulate systemic immune cell fate at homeostasis.
Materials and methods
Mice
Six- to 8-week-old wild-type (WT) C57BL/6 mice were obtained from Jackson Laboratories and maintained in strict accordance with a protocol approved by the University of Pennsylvania Institutional Animal Care and Use Committee. Congenic Nod1−/−, Tlr4−/−, and Nod2Tlr2−/− mice were described previously.27 Germ-free C57BL/6 mice were obtained from the University of Pennsylvania Gnotobiotic Mouse Facility. All germ-free mice were 8-week-old females, age- and sex-matched to conventional controls. All mice were euthanized by CO2 inhalation followed by cardiac puncture.
Antibiotic treatment
All antibiotics were administered in drinking water ad libitum. Mice received sterile-filtered tap water supplemented with either a broad-spectrum antibiotic cocktail (1 g/L each of neomycin [Fisher], vancomycin [Santa Cruz], metronidazole [Sigma-Aldrich], ampicillin [Corning], and gentamicin [Life Technologies]), 1 g/L of neomycin or metronidazole alone, or conventional tap water. Water was changed every 3 to 4 days, and the treatment regimen was maintained for 3 to 4 weeks.
Tissue processing and cell isolation
Bone marrow parenchyma was flushed from the femora of mice with RPMI 1640 (Corning) supplemented with 5% fetal bovine serum (FBS) (GIBCO). Murine blood was obtained from retro-orbital sinuses following isoflurane anesthesia (Santa Cruz) or by cardiac puncture. Blood samples were anticoagulated with 0.1 M EDTA (Invitrogen), and erythrocytes were lysed with ACK lysis buffer (Life Technologies) for 5 minutes at room temperature. Splenocytes were isolated by homogenization over 40-µm nylon cell strainers (Falcon) into RPMI 1640 + 5% FBS. Murine ilea were processed as described previously,28 and 40-µm nylon cell strainers (Falcon) were used to obtain intestinal cell homogenates before stimulation and flow cytometric analysis.
Flow cytometry
After resuspension of cells in phosphate-buffered saline (PBS) with 1% bovine serum albumin, Fc receptor blocking was performed with 1:100 dilution of anti-CD16/32 (BD Biosciences, 2.4G2). Cellular viability/integrity was assessed by Fixable Viability Dye eFluor780 (eBioscience). Surface marker staining was performed for 30 minutes at 4°C, antibodies diluted 1:150, against: CD45 (eBioscience, 30-F11); CD11b (M1/70), CD11c (N418), Ly6G (1A8), Ly6C (AL-21), CXCR4 (L276F12), CD62L (MEL-14), F4/80 (BM8), CD4 (GK1.5), CD8 (53-6.7), B220 (RA3-B26), T-cell receptor (TCR) αβ (H57-597), and TCRγδ (GL3) (all from BioLegend). Stimulation of IL-17A production was performed via incubation of murine ileal cells with 50 ng/mL phorbol 12-myristate-13-acetate (Sigma-Aldrich) and 750 ng/mL ionomycin (Sigma-Aldrich) for 6 hours. Intracellular staining for IL-17A (BioLegend, TC11-18H10.1, 1:50) and RORγt (eBioscience, B2D, 1:50) was performed after fixation and permeabilization with BD Cytofix/Cytoperm as per manufacturer’s instructions and treatment with 1:1000 GolgiStop (BD) as described previously.28 All cytometry was performed on a BD LSRII flow cytometer and analyzed using FlowJo software (Tree Star).
Ex vivo apoptosis assays
We applied freshly obtained, erythrocyte-lysed blood and cells obtained from splenic or bone marrow parenchyma to 96-well plates, with ∼105 cells and 200 μL RPMI + 5% FBS per well. No antibiotics or cytokines were added to ex vivo culture media. Plates were incubated at 37°C and removed at the indicated time points before examination by flow cytometry. Apoptosis among each cell population gated during analysis was measured by staining with Annexin V (1:20) in Annexin V Binding Buffer (BioLegend) for 15 minutes at room temperature. Viable cells were identified by exclusion of Annexin V Fixable Viability Stain.
BrdU pulse-chase assays
The 5-bromo-2′-deoxyuridine (BrdU) incorporation was measured as described previously29 with the following modifications: Mice were given 3 intraperitoneal (IP) injections of 2 mg BrdU (Life Technologies), spaced 3 hours apart, in 100 μL PBS. After leukocytes were obtained by retro-orbital bleeding (∼100 µL) or isolation of splenic or bone marrow parenchyma, BrdU incorporation was measured by flow cytometry (BioLegend, Bu20a, 1:20) at the indicated time points postinjection after fixation and permeabilization per manufacturer’s instructions. Decreases in the proportion of BrdU+ cells were fit to a 1-phase exponential decay equation, N(t) = N(0)e−γt, where N represents the BrdU+ fraction over time t and the rate constant γ is calculated.
Competitive adoptive transfer assays
Mature murine bone marrow neutrophils were adoptively transferred into the bloodstreams of recipient mice as described previously,30 with the following modifications: Neutrophils obtained from WT or Nod1−/− mice (∼5 × 106 cells per mL) were stained for 30 minutes at 37°C with 5 µM of either carboxyfluorescein diacetate succinimidyl ester (CFSE) or Cell Proliferation Dye eFluor 450 (eF450, eBioscience). After washing twice and resuspension in PBS, 2.5 × 106 neutrophils of each genotype were combined 1:1 before retro-orbital injection into recipient mice (200 µL, 25 × 106 cells per mL). Peripheral blood was collected 4 hours after transfer, and relative abundance of each neutrophil population was assessed by flow cytometry. Dye-switch control experiments were performed as indicated.
16S rDNA collection, quantification, and sequencing
Stool samples were collected from neomycin-treated and control mice on days 7, 14, and 21 after initiation of antibiotic treatment, with ileum contents also collected on day 21. Samples were flash frozen at −80°C, and DNA was extracted using Zymo ZR Fecal Miniprep Kits. 16S ribosomal DNA (rDNA) copy number was quantified by quantitative polymerase chain reaction (PCR) relative to a standard curve constructed from a TOPO vector containing Escherichia coli 16S rDNA. PCR reactions were performed in triplicate using the following 16S rDNA-specific primers: forward, 5′-AGAGTTTGATCCTGGCTCAG-3′; reverse, 5′-CTGCTGCCTYCCGTA-3′; probe, 5′-FAM-TA+ACA+CATG+CA+AGTC+GA/3BHQ1-3′ (“+” precedes position of LNA base). 16S rDNA metagenomic sequencing was performed as described previously on the Illumina HiSeq platform, using bar-coded primers aligned to the V1V2 region.31 Data were analyzed and quality controlled using the QIIME pipeline v.1.8, and phylogenetic trees were constructed using FastTree v.2.1.3 with default parameters.32,33
Ligand treatments and cytokine neutralization
To stimulate Nod1 signaling in vivo, we injected mice IP with 2 doses of 100 µg C12-iE-DAP (InvivoGen), a well-described, acylated derivative of the Nod1 dipeptide ligand iE-DAP. We used C12-iE-DAP because it bears the minimal requisite peptidoglycan structure necessary for specific Nod1 stimulation and acylation enhances distribution into lipophilic tissue environments.34 Injections were given 12 and 2 hours before euthanasia for ex vivo apoptosis assays and at 48 and 72 hours after BrdU injections for in vivo myeloid cell turnover experiments. C12-iE-DAP (100 µg) was administered 12, 48, and 72 hours prior to euthanasia for intestinal flow cytometry assays. Neutralization of IL-17A was achieved through 2 IP injections of 100 µg anti-IL-17A monoclonal antibody (BioXCell, 17F3) or immunoglobulin (Ig) G1 isotype control antibody (MOPC-21), performed at 12 and 2 hours before euthanasia for ex vivo apoptosis assays and at 48 and 72 hours after BrdU injections for in vivo myeloid cell turnover experiments.
Statistics
Data are displayed as mean ± standard error of the mean (SEM) except in scatter plots, where horizontal bars depict median values. Except where indicated otherwise, statistical significance was assessed by 2-tailed Student t test for pairwise comparisons and analysis of variance (ANOVA) with Newman-Keuls posttest for comparisons of >2 groups. For all analyses, P < .05 was considered statistically significant.
Results
Antibiotic treatment accelerates turnover of circulating phagocytes from the bloodstream
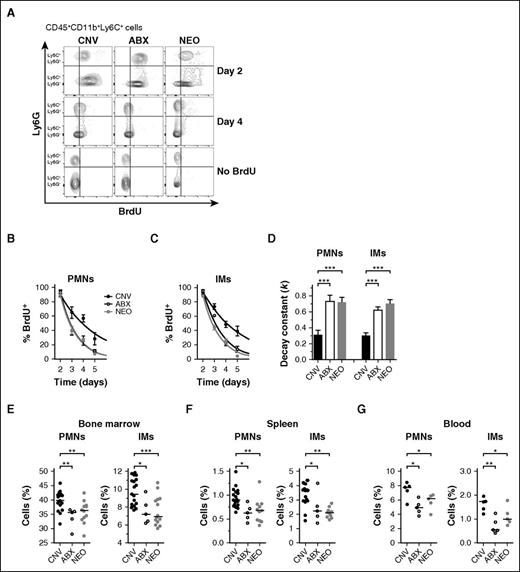
To assess whether the microbiota influences the circulating longevity of blood phagocytes, we performed BrdU pulse-chase assays in mice treated for 4 weeks with conventional tap water or broad spectrum antibiotics. After pulsing mice IP with BrdU, we measured the fraction of BrdU+ neutrophils (PMNs) and IMs in the bloodstream over time (Figure 1A; supplemental Figure 1, available on the Blood Web site). Previous studies have shown that BrdU is not mitogenic at these doses, is incorporated most readily into the numerous late precursors giving rise to mature phagocytes in the bone marrow, and that circulating phagocytes reach saturation with BrdU ∼2 days after its administration.29,35 Accordingly, we found that nearly 100% of bloodstream PMNs (Figure 1B) and IMs (Figure 1C) were BrdU+ at 2 days postinjection. This saturation occurred irrespective of antibiotic treatment. Next, we tracked the contraction of BrdU+ cells from the blood of antibiotic-treated and control mice over the following 3 days. In contrast to the equivalent frequencies of labeled cells at day 2, treatment with broad-spectrum antibiotics significantly accelerated the elimination of BrdU+ PMNs and IMs from circulation (Figure 1B-C), portrayed quantitatively by enhanced decay kinetics when set to 1-phase exponential decay equations (Figure 1D). BrdU retention in splenic PMNs and IMs mirrored those from the bloodstream (supplemental Figure 2A).
Antibiotic treatment accelerates turnover of circulating phagocytes from the bloodstream. (A) Representative flow cytometry plots from BrdU pulse-chase experiments using mice previously treated with conventional water (CNV, black), broad-spectrum antibiotics (ABX, open), or neomycin (NEO, gray). Events represent CD45+CD11b+Ly6C+ cells stained for intracellular BrdU content and Ly6G (neutrophils: Ly6G+; inflammatory monocytes: Ly6G−) on days 2 and 4 after BrdU injection, compared with no-BrdU controls. Frequency of BrdU+ events among blood neutrophils (PMNs, B) and IMs (C) assessed on days 2 through 5 after BrdU administration. (D) Rate constants (k, day−1) for 1-phase exponential decay quantified from BrdU+ frequencies depicted in panels B and C. Steady-state cell frequencies (% of CD45+ events) of PMNs and IMs measured from the bone marrow (E), spleens (F), and blood (G) of CNV-, ABX-, and NEO-treated mice. Absolute cell numbers: Bone marrow: CNV PMNs 9.99 ± 0.19, ABX PMNs 8.51 ± 0.39, NEO PMNs 8.98 ± 0.29, CNV IMs 2.05 ± 0.07, ABX IMs 1.60 ± 0.14, NEO IMs 1.61 ± 0.09 (all ×106 cells per femur). Spleen: CNV PMNs 1.01 ± 0.05, ABX PMNs 0.67 ± 0.08, NEO PMNs 0.72 ± 0.08, CNV IMs 3.26 ± 0.20, ABX IMs 2.40 ± 0.46, NEO IMs 2.07 ± 0.11 (all ×106 cells per spleen). Blood: CNV PMNs 882 ± 64, ABX PMNs 472 ± 38, NEO PMNs 530 ± 45, CNV IMs 227 ± 18, ABX IMs 92 ± 22, NEO IMs 154 ± 14 (all cells per μL). Horizontal lines in scatter plots depict median values. All other data presented as mean ± SEM with ≥5 mice per group. Statistical significance was assessed by Student t test for pairwise comparisons and 1-way ANOVA with Newman-Keuls posttest for comparisons of >2 conditions. *P < .05; **P < .01; ***P < .001. NS, not significant.
Antibiotic treatment accelerates turnover of circulating phagocytes from the bloodstream. (A) Representative flow cytometry plots from BrdU pulse-chase experiments using mice previously treated with conventional water (CNV, black), broad-spectrum antibiotics (ABX, open), or neomycin (NEO, gray). Events represent CD45+CD11b+Ly6C+ cells stained for intracellular BrdU content and Ly6G (neutrophils: Ly6G+; inflammatory monocytes: Ly6G−) on days 2 and 4 after BrdU injection, compared with no-BrdU controls. Frequency of BrdU+ events among blood neutrophils (PMNs, B) and IMs (C) assessed on days 2 through 5 after BrdU administration. (D) Rate constants (k, day−1) for 1-phase exponential decay quantified from BrdU+ frequencies depicted in panels B and C. Steady-state cell frequencies (% of CD45+ events) of PMNs and IMs measured from the bone marrow (E), spleens (F), and blood (G) of CNV-, ABX-, and NEO-treated mice. Absolute cell numbers: Bone marrow: CNV PMNs 9.99 ± 0.19, ABX PMNs 8.51 ± 0.39, NEO PMNs 8.98 ± 0.29, CNV IMs 2.05 ± 0.07, ABX IMs 1.60 ± 0.14, NEO IMs 1.61 ± 0.09 (all ×106 cells per femur). Spleen: CNV PMNs 1.01 ± 0.05, ABX PMNs 0.67 ± 0.08, NEO PMNs 0.72 ± 0.08, CNV IMs 3.26 ± 0.20, ABX IMs 2.40 ± 0.46, NEO IMs 2.07 ± 0.11 (all ×106 cells per spleen). Blood: CNV PMNs 882 ± 64, ABX PMNs 472 ± 38, NEO PMNs 530 ± 45, CNV IMs 227 ± 18, ABX IMs 92 ± 22, NEO IMs 154 ± 14 (all cells per μL). Horizontal lines in scatter plots depict median values. All other data presented as mean ± SEM with ≥5 mice per group. Statistical significance was assessed by Student t test for pairwise comparisons and 1-way ANOVA with Newman-Keuls posttest for comparisons of >2 conditions. *P < .05; **P < .01; ***P < .001. NS, not significant.
Surprisingly, treating mice with the aminoglycoside neomycin alone was sufficient to reproduce the impact of broad-spectrum antibiotics on the longevity of both PMNs and IMs (Figure 1A-D). This corresponded with modest but significant decreases in the frequencies of total PMNs and IMs in the bone marrow (Figure 1E), spleens (Figure 1F), and blood (Figure 1G) of mice treated with broad-spectrum antibiotics or neomycin, comparable to differences observed previously with broad-spectrum antibiotics and germ-free mice.23,28 Taken together, these data provide evidence that the neomycin-sensitive gut microbiota augments the circulating lifespan of bloodstream phagocytes in vivo.
Antibiotic treatment leads to impaired phagocyte survival ex vivo
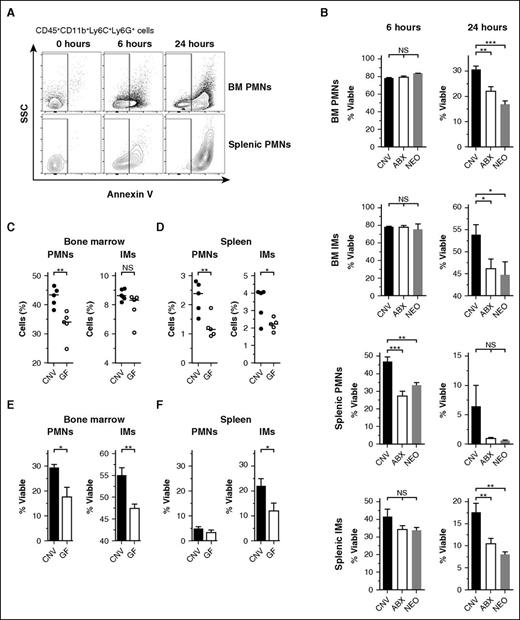
Following exit from the bloodstream, senescent phagocytes undergo spontaneous apoptosis before clearance by tissue-resident macrophages.5,36 We asked whether the microbiota influences the duration of phagocyte viability and resistance to spontaneous cell death. Because apoptotic cells are cleared immediately in vivo and do not accumulate at steady state,37 we determined the lifespan of PMNs and IMs ex vivo in cultures lacking growth factors or cytokines. At 6 or 24 hours after harvest of bone marrow or splenic parenchyma, we measured the proportion of phagocytes that remained viable and excluded Annexin V, a stain that detects early apoptosis by binding surface-exposed phosphatidylserine (Figure 2A, representative gating for neutrophils).
Antibiotic treatment corresponds with impaired phagocyte survival ex vivo. (A) Representative flow cytometry plots of neutrophils (PMNs, gated on CD45+CD11b+Ly6C+Ly6G+) obtained from bone marrow (BM) and spleens and allowed to incubate ex vivo for 0, 6, or 24 hours before staining with Annexin V. Nonapoptotic, viable cells were quantified by gating on Annexin V− events (SSC, side scatter). (B) Assessment of Annexin V− event frequency (% Viable) among PMNs and IMs obtained from the BM and spleens of conventional (CNV, black), broad-spectrum antibiotic-treated (ABX, open), and neomycin-treated (NEO, gray) mice following 6 or 24 hours incubation ex vivo. Steady-state frequencies (% of CD45+ events) of PMNs and IMs obtained from the BM (C) and spleens (D) of CNV (black circles) and germ-free mice (GF, open circles). Absolute cell numbers: BM: CNV PMNs 10.83 ± 0.37, GF PMNs 8.40 ± 0.56, CNV IMs 2.24 ± 0.04, GF IMs 2.01 ± 0.12 (all ×106 cells per femur). Spleen: CNV PMNs 1.07 ± 0.12, GF PMNs 0.58 ± 0.08, CNV IMs 2.67 ± 0.33, GF IMs 1.81 ± 0.15 (all ×106 cells per spleen). Survival ex vivo of PMNs and IMs from the BM (E, 24 hours) and spleens (F, 6 hours) of CNV and GF mice. Horizontal lines in scatter plots depict median values. All other data presented as mean ± SEM with ≥3 to 5 mice per group. Statistical significance was assessed by Student t test for pairwise comparisons and 1-way ANOVA with Newman-Keuls posttest for comparisons of >2 conditions. *P < .05; **P < .01; ***P < .001.
Antibiotic treatment corresponds with impaired phagocyte survival ex vivo. (A) Representative flow cytometry plots of neutrophils (PMNs, gated on CD45+CD11b+Ly6C+Ly6G+) obtained from bone marrow (BM) and spleens and allowed to incubate ex vivo for 0, 6, or 24 hours before staining with Annexin V. Nonapoptotic, viable cells were quantified by gating on Annexin V− events (SSC, side scatter). (B) Assessment of Annexin V− event frequency (% Viable) among PMNs and IMs obtained from the BM and spleens of conventional (CNV, black), broad-spectrum antibiotic-treated (ABX, open), and neomycin-treated (NEO, gray) mice following 6 or 24 hours incubation ex vivo. Steady-state frequencies (% of CD45+ events) of PMNs and IMs obtained from the BM (C) and spleens (D) of CNV (black circles) and germ-free mice (GF, open circles). Absolute cell numbers: BM: CNV PMNs 10.83 ± 0.37, GF PMNs 8.40 ± 0.56, CNV IMs 2.24 ± 0.04, GF IMs 2.01 ± 0.12 (all ×106 cells per femur). Spleen: CNV PMNs 1.07 ± 0.12, GF PMNs 0.58 ± 0.08, CNV IMs 2.67 ± 0.33, GF IMs 1.81 ± 0.15 (all ×106 cells per spleen). Survival ex vivo of PMNs and IMs from the BM (E, 24 hours) and spleens (F, 6 hours) of CNV and GF mice. Horizontal lines in scatter plots depict median values. All other data presented as mean ± SEM with ≥3 to 5 mice per group. Statistical significance was assessed by Student t test for pairwise comparisons and 1-way ANOVA with Newman-Keuls posttest for comparisons of >2 conditions. *P < .05; **P < .01; ***P < .001.
In concordance with accelerated turnover from the bloodstream, PMNs and IMs from mice treated with broad-spectrum antibiotics or neomycin entered apoptosis more rapidly than controls (Figure 2B). Accelerated cell death was apparent at 6 hours postharvest among splenic PMNs and at 24 hours among phagocytes from both the bone marrow and spleen. Notably, neomycin treatment had no effect on the abundance of CD62LloCXCR4hi “aged” PMNs isolated from the bloodstream (supplemental Figure 3). Apoptosis among PMNs and IMs isolated from the bloodstream mirrored that of splenic phagocytes at 6 hours (supplemental Figure 4A). In contrast, neomycin treatment had no measurable impact on the apoptosis of splenic lymphocytes, fibroblasts, or endothelial cells (supplemental Figure 4B). Splenic macrophages exhibited apoptosis kinetics similar to that of IMs (supplemental Figure 4C), likely reflecting the monocytic origin of some of these cells.38
To verify that the depletion of microbes was responsible for accelerated phagocyte apoptosis rather than a direct effect of antibiotics, we repeated the ex vivo apoptosis assays using germ-free mice. Similar to the effects observed upon treatment with antibiotics, germ-free mice trended toward lower frequencies of PMNs and IMs in the bone marrow (Figure 2C) and spleen (Figure 2D) and exhibited accelerated apoptosis when incubated ex vivo (Figure 2E-F). In contrast, treating mice solely with metronidazole, which preferentially targets anaerobes,39 had no impact on phagocyte viability (data not shown). Thus, a distinct, neomycin-sensitive cohort of microbes regulates the cellular lifespan of mature phagocytes in addition to their turnover from the bloodstream.
Treatment with neomycin stably alters the community structure of the gut microbiota
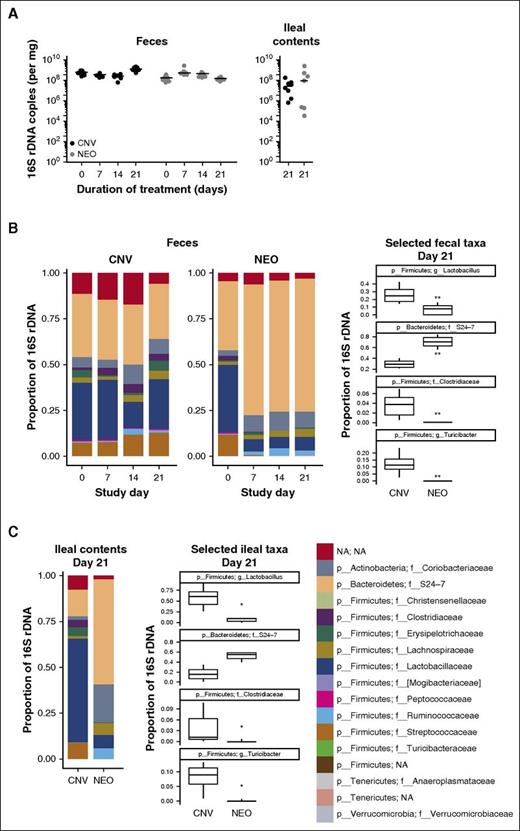
Treatment with neomycin recapitulated the impact of broad-spectrum antibiotics on phagocyte turnover. Thus, we sought next to characterize the impact of this gut-restricted aminoglycoside on the murine intestinal microbiota.40,41 After collecting weekly fecal samples and harvesting ileal contents 21 days after initiation of treatment, we extracted DNA and performed 16S rDNA quantitative PCR to quantify changes in intestinal microbial abundance. In contrast to reductions described previously with broad-spectrum antibiotics,42 we observed no significant differences in fecal or ileal 16S rDNA copy number between neomycin-treated and conventional mice (Figure 3A). However, metagenomic sequencing of intestinal contents revealed stable and reproducible shifts in microbial community architecture at both sites within 1 week after the onset of treatment (Figure 3B-C). Notably, Lactobacillaceae family bacteria dropped from ∼25% of fecal microbes among conventional mice to <10% with neomycin, paired with a ∼40% increase in anaerobic S24-7-family Bacteroidetes species (Figure 3B). This redistribution concurs with previous reports showing sensitivity among lactobacilli and resistance among anaerobes to aminoglycosides in vitro.43,44
Metagenomic analysis of the murine intestinal microbiota upon treatment with neomycin. (A) Quantitative real-time PCR analysis of 16S rDNA copy number from the feces or ileal contents of mice treated with conventional water (CNV, black) or neomycin (NEO, gray). Horizontal lines depict median values. (B) Family-level phylogenetic analyses of fecal bacterial sequences from conventional and neomycin-treated mice. Frequencies of significantly altered bacterial taxa in feces were compared individually at day 21. (C) Family-level phylogenetic analysis of ileal contents taken at day 21, with individual frequency comparisons of significantly altered taxa. All data represent averages from 8 mice in each treatment condition, housed as pairs in 4 separate cages. Box-and-whisker plots depict medians ± interquartile range. Statistical significance for all comparisons was analyzed by Kruskal-Wallis tests with correction for false discovery rate. *P < .05; **P < .01.
Metagenomic analysis of the murine intestinal microbiota upon treatment with neomycin. (A) Quantitative real-time PCR analysis of 16S rDNA copy number from the feces or ileal contents of mice treated with conventional water (CNV, black) or neomycin (NEO, gray). Horizontal lines depict median values. (B) Family-level phylogenetic analyses of fecal bacterial sequences from conventional and neomycin-treated mice. Frequencies of significantly altered bacterial taxa in feces were compared individually at day 21. (C) Family-level phylogenetic analysis of ileal contents taken at day 21, with individual frequency comparisons of significantly altered taxa. All data represent averages from 8 mice in each treatment condition, housed as pairs in 4 separate cages. Box-and-whisker plots depict medians ± interquartile range. Statistical significance for all comparisons was analyzed by Kruskal-Wallis tests with correction for false discovery rate. *P < .05; **P < .01.
The ileal microbiota exhibited changes broadly similar to those observed in fecal samples (Figure 3C). However, given the previously described immunoregulatory role of segmented filamentous bacteria (SFB) residing within the small intestine,45 we queried directly whether neomycin induced changes in the abundance of SFB species (eg, Candidatus Arthromitus). We detected no SFBs in control or treated mice, a finding consistent with previous analyses of mice acquired from Jackson Laboratories.46 Although the effects of antibiotics on the intestinal microbial ecosystem are complex, these results suggest that the impact of neomycin on phagocyte lifespan corresponds with a discrete and stable signature of altered commensal flora. However, the nature of the microbial signal connecting gut commensals to phagocyte homeostasis remained unclear.
Nod1 mediates the microbial influence on phagocyte lifespan at homeostasis
We previously described a role for the intracellular peptidoglycan sensor Nod1 in driving microbiota-mediated stimulation of neutrophil phagocytic capacity ex vivo.27 Because the peptidoglycan of members of the Lactobacillaceae family depleted by neomycin treatment bear iE-DAP and stimulate Nod1 in vitro,47,48 we hypothesized that the microbiota stimulates this cytoplasmic sensor to regulate phagocyte homeostasis in the absence of infection. Previous work has demonstrated a minimal impact of Nod1 gene deletion on the composition of the murine microbiome at homeostasis.49 To determine whether Nod1 signaling instead stimulates the viability of PMNs and IMs, we repeated ex vivo phagocyte apoptosis assays using Nod1−/− mice.
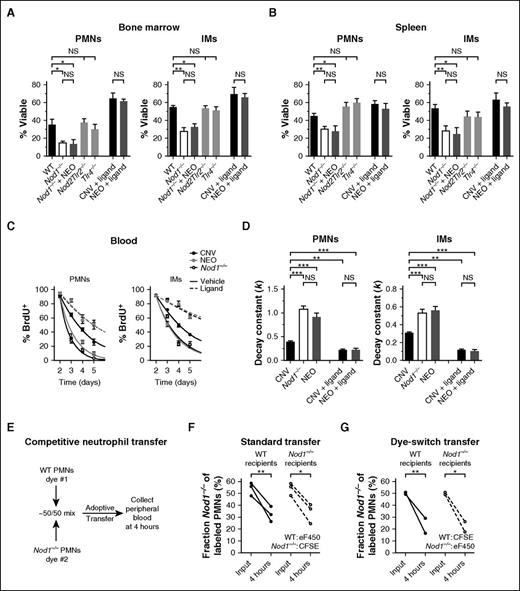
PMNs and IMs from both the bone marrow (Figure 4A) and spleens (Figure 4B) of Nod1−/− mice entered apoptosis significantly more rapidly than those of WT controls, instead resembling phagocytes from neomycin-treated mice. Treating Nod1−/− mice with neomycin produced no additional increase in the kinetics of phagocyte apoptosis, reinforcing that Nod1 is responsible for relaying the stimulatory signal from the flora into changes in cellular viability. In contrast, the lifespan of phagocytes from Nod2Tlr2−/− and Tlr4−/− mice resembled those of WT controls despite well-described roles for these sensors in regulating granulopoiesis and phagocyte activation.22,50,51 To assess whether signaling through Nod1 was sufficient to recapitulate the impact of the microbiota on cellular lifespan, we repeated the apoptosis assays after injecting mice IP with the acylated Nod1 ligand C12-iE-DAP. Ligand treatment rescued the viability of phagocytes from neomycin-treated mice such that their lifespan was indistinguishable from conventional mice while conferring only a modest survival benefit to the conventional cells.
The intracellular peptidoglycan sensor Nod1 is necessary and sufficient to mediate the microbial influence on phagocyte lifespan. Quantification of cell survival ex vivo (Annexin V−, % Viable) among neutrophils (PMNs) and IMs from the bone marrow (A, 24 hours) and spleens (B, 6 hours) of conventional WT, Nod1−/−, NEO-treated Nod1−/−, Nod2Tlr2−/−, and Tlr4−/− mice. Cellular lifespan was measured similarly for phagocytes obtained from CNV- and NEO-treated mice after IP injection of the Nod1 ligand C12-iE-DAP (Ligand), with 100 μg doses given 12 and 2 hours before euthanasia. (C) BrdU pulse-chase assays. BrdU+ frequency among PMNs and IMs from the bloodstream of CNV (black), NEO (gray), and Nod1−/− mice (open), measured on days 2 to 5 after systemic BrdU administration. Mice received injections of vehicle (1% dimethyl sulfoxide in PBS, solid curves) or 100 μg C12-iE-DAP (Ligand, hashed curves) on days 2 and 3 after injection of BrdU. (D) Rate constants (k, day−1) for 1-phase exponential decay quantified from BrdU+ frequencies depicted in panel C. Data presented as mean ± SEM with ≥5 mice per group. (E) Schematic for competitive neutrophil adoptive transfer assay. (F) Competitive adoptive transfer of eF450-labeled WT PMNs and CFSE-labeled Nod1−/− PMNs into WT (black circles) or Nod1−/− (open circles) recipient mice. Data depict the fraction Nod1−/− among labeled PMNs 4 hours after standard transfer. (G) Competitive adoptive transfer of CFSE-labeled WT PMNs and eF450-labeled Nod1−/− PMNs (dye-switch control). Statistical significance was assessed by Student t test for pairwise comparisons and 1-way ANOVA with Newman-Keuls posttest for comparisons of >2 conditions. *P < .05; **P < .01; ***P < .001.
The intracellular peptidoglycan sensor Nod1 is necessary and sufficient to mediate the microbial influence on phagocyte lifespan. Quantification of cell survival ex vivo (Annexin V−, % Viable) among neutrophils (PMNs) and IMs from the bone marrow (A, 24 hours) and spleens (B, 6 hours) of conventional WT, Nod1−/−, NEO-treated Nod1−/−, Nod2Tlr2−/−, and Tlr4−/− mice. Cellular lifespan was measured similarly for phagocytes obtained from CNV- and NEO-treated mice after IP injection of the Nod1 ligand C12-iE-DAP (Ligand), with 100 μg doses given 12 and 2 hours before euthanasia. (C) BrdU pulse-chase assays. BrdU+ frequency among PMNs and IMs from the bloodstream of CNV (black), NEO (gray), and Nod1−/− mice (open), measured on days 2 to 5 after systemic BrdU administration. Mice received injections of vehicle (1% dimethyl sulfoxide in PBS, solid curves) or 100 μg C12-iE-DAP (Ligand, hashed curves) on days 2 and 3 after injection of BrdU. (D) Rate constants (k, day−1) for 1-phase exponential decay quantified from BrdU+ frequencies depicted in panel C. Data presented as mean ± SEM with ≥5 mice per group. (E) Schematic for competitive neutrophil adoptive transfer assay. (F) Competitive adoptive transfer of eF450-labeled WT PMNs and CFSE-labeled Nod1−/− PMNs into WT (black circles) or Nod1−/− (open circles) recipient mice. Data depict the fraction Nod1−/− among labeled PMNs 4 hours after standard transfer. (G) Competitive adoptive transfer of CFSE-labeled WT PMNs and eF450-labeled Nod1−/− PMNs (dye-switch control). Statistical significance was assessed by Student t test for pairwise comparisons and 1-way ANOVA with Newman-Keuls posttest for comparisons of >2 conditions. *P < .05; **P < .01; ***P < .001.
To ascertain whether Nod1 signaling plays a similarly central role in regulating phagocyte turnover in vivo, we subjected Nod1−/− mice to BrdU pulse-chase assays (Figure 4C). Congruent with their impaired survival ex vivo, PMNs and IMs disappeared from circulation more quickly in knockout mice than in controls and knockout phagocyte turnover kinetics were indistinguishable from that of neomycin-treated animals (Figure 4D). Further, administration of C12-iE-DAP markedly delayed turnover of both PMNs and IMs from the blood and equalized the longevity of phagocytes from conventional and neomycin-treated mice (Figure 4C-D). Treatment with C12-iE-DAP had no effect on the longevity of PMNs and IMs from Nod1−/− mice (supplemental Figure 2B).
Finally, we performed competitive adoptive transfer experiments using 1:1 mixtures of WT and Nod1−/− PMNs to assess whether Nod1-mediated stimulation of cell longevity persists when phagocytes are transplanted into genotype-discordant host environments (Figure 4E). IMs were not adoptively transferred because of significantly lower availability of donor cells in the murine bone marrow. PMNs transferred from Nod1−/− donors were disproportionately cleared from the recipient bloodstream by 4 hours posttransfer when compared with WT cells (Figure 4F). This rapid turnover persisted in both WT and Nod1−/− recipients and was unaffected by the dyes used to label each donor neutrophil population (Figure 4G). Collectively, these data reveal that recognition of iE-DAP by Nod1 is both necessary and sufficient to mediate the stimulatory impact of the flora on phagocyte lifespan.
IL-17A relays Nod1-dependent microbial signals to promote phagocyte longevity
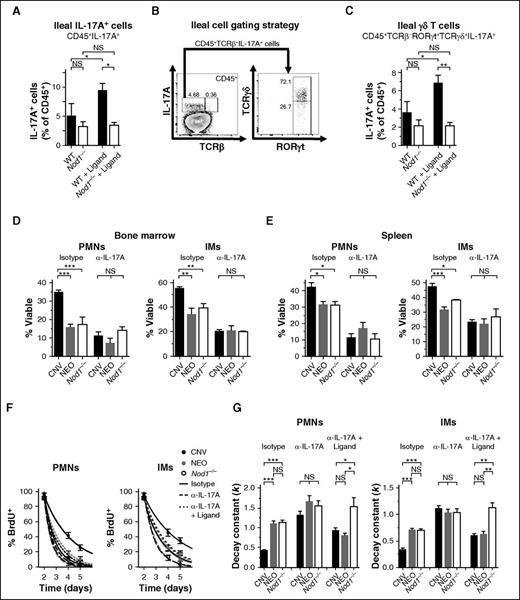
Although Nod1-mediated sensing of peptidoglycan from the microbiota is essential for the maintenance of gut mucosal immunity,52 how these signals are transmitted to instruct systemic immune homeostasis remains poorly understood. Nod1 signaling drives systemic responses to infection primarily through the liberation of cytokines that recruit and activate phagocytes.53 We predicted that similar signaling regulates phagocyte lifespan in the steady state. We focused our studies on IL-17A, a key cytokine orchestrating myeloid cell development, function, and survival,28,54,55 because its production has been shown to be stimulated by both the gut microbiota and Nod1 signaling.46,56 Although we observed a trend toward fewer IL-17A-producing leukocytes in the ilea of Nod1−/− mice compared with WT mice, administration of C12-iE-DAP significantly expanded the number of IL-17A+ cells from the ilea of WT mice and exerted no effect in Nod1−/− animals (Figure 5A). The majority of IL-17A+ ileal cells did not express TCRαβ (>90%), with a large representation of γδ T cells (Figure 5B). Accordingly, treatment of WT mice with C12-iE-DAP significantly expanded the pool of ileal IL-17A+ γδ T cells, whereas no such expansion occurred in Nod1−/− mice (Figure 5C). These results indicate that Nod1 signaling is sufficient to stimulate the liberation of IL-17A from the murine intestine.
IL-17A relays Nod1-dependent microbial signals to promote phagocyte longevity. (A) Frequency of IL-17A-producing CD45+ cells from the ilea of WT and Nod1−/− mice after 3 injections of 100 µg C12-iE-DAP (Ligand) or vehicle control. (B) Gating strategy for analysis of IL-17A-producing ileal cells (WT example shown). (C) Quantification of IL-17A-producing ileal γδ T cells following treatment as in panel A. (D) Assessment of cellular lifespan ex vivo (Annexin V−, % Viable) among neutrophils (PMNs) and IMs. Survival of cells from the bone marrow (D, 24 hours) and spleens (E, 6 hours) was quantified for WT, neomycin-treated WT, and Nod1−/− mice after 100 µg injections of IgG1 isotype control (Isotype) or neutralizing anti-IL-17A antibody (α-IL-17A), given 12 and 2 hours before euthanasia. (F) BrdU pulse-chase assays. Quantification of BrdU+ frequency among PMNs and IMs from the bloodstream of CNV (black), NEO (gray), and Nod1−/− mice (open). Mice received injections of 100 μg of IgG1 isotype control (solid curves) or IL-17A-neutralizing antibody (hashed curves) on days 2 and 3 after administration of BrdU, with or without concurrent injections of 100 µg C12-iE-DAP (Ligand). BrdU+ frequency was assessed on days 2, 4, and 5 after BrdU injection. (G) Rate constants (k, day−1) for 1-phase exponential decay quantified from BrdU+ frequencies depicted in (F). All data presented as mean ± SEM with ≥4 mice per group. Statistical significance was assessed by Student t test for pairwise comparisons and 1-way ANOVA with Newman-Keuls posttest for comparisons of >2 conditions. *P < .05; **P < .01; ***P < .001.
IL-17A relays Nod1-dependent microbial signals to promote phagocyte longevity. (A) Frequency of IL-17A-producing CD45+ cells from the ilea of WT and Nod1−/− mice after 3 injections of 100 µg C12-iE-DAP (Ligand) or vehicle control. (B) Gating strategy for analysis of IL-17A-producing ileal cells (WT example shown). (C) Quantification of IL-17A-producing ileal γδ T cells following treatment as in panel A. (D) Assessment of cellular lifespan ex vivo (Annexin V−, % Viable) among neutrophils (PMNs) and IMs. Survival of cells from the bone marrow (D, 24 hours) and spleens (E, 6 hours) was quantified for WT, neomycin-treated WT, and Nod1−/− mice after 100 µg injections of IgG1 isotype control (Isotype) or neutralizing anti-IL-17A antibody (α-IL-17A), given 12 and 2 hours before euthanasia. (F) BrdU pulse-chase assays. Quantification of BrdU+ frequency among PMNs and IMs from the bloodstream of CNV (black), NEO (gray), and Nod1−/− mice (open). Mice received injections of 100 μg of IgG1 isotype control (solid curves) or IL-17A-neutralizing antibody (hashed curves) on days 2 and 3 after administration of BrdU, with or without concurrent injections of 100 µg C12-iE-DAP (Ligand). BrdU+ frequency was assessed on days 2, 4, and 5 after BrdU injection. (G) Rate constants (k, day−1) for 1-phase exponential decay quantified from BrdU+ frequencies depicted in (F). All data presented as mean ± SEM with ≥4 mice per group. Statistical significance was assessed by Student t test for pairwise comparisons and 1-way ANOVA with Newman-Keuls posttest for comparisons of >2 conditions. *P < .05; **P < .01; ***P < .001.
Neutralization of IL-17A markedly impaired the survival ex vivo of PMNs and IMs from the bone marrow (Figure 5D) and spleens (Figure 5E) of conventionally treated mice. The blockade conferred limited impact on cells from neomycin-treated or Nod1−/− animals, resulting in equal cellular viability across all groups in the absence of IL-17A signaling. BrdU pulse-chase assays revealed similar results in vivo, with neutralization of IL-17A phenocopying neomycin treatment or Nod1 loss in accelerating the turnover of PMNs and IMs from the bloodstream (Figure 5F-G). Furthermore, simultaneous treatment with C12-iE-DAP and IL-17A neutralization significantly reduced the impact of Nod1 stimulation on the lifespans of PMNs and IMs (P < .001) and abrogated the effect of the microbiota on phagocyte longevity (Figure 5F-G). Collectively, these results demonstrate that neomycin-sensitive microbes, Nod1, and IL-17A comprise requisite components of a common signaling axis that governs systemic phagocyte lifespan at homeostasis.
Discussion
Cell turnover is a key component of immune homeostasis, and accumulating evidence suggests that failure to regulate the lifespan of circulating phagocytes can impair inflammatory responses and host defense. Here, we showed that the gut microbiota governs the viability and circulating lifespan of PMNs and IMs in the steady state. Our observations highlight how broad-spectrum antibiotic therapy may negatively impact systemic myeloid homeostasis and reveal a signaling axis through which commensal microbes promote the availability of circulating phagocytes. As disruptions of commensal microbial communities are associated with a growing set of inflammatory, infectious, and neoplastic diseases, mechanistic understanding of the microbial influence on innate immunity during health may reveal therapeutic targets when this relationship is disturbed.57,58
We found that treating mice with broad-spectrum antibiotics accelerated the apoptotic cell death and clearance of circulating neutrophils, comprising 2 essential steps in phagocyte turnover. Several previous studies have documented effects of the microbiota on distinct aspects of neutrophil biology, including stimulation of neutrophil development in the bone marrow,22-24 trafficking to inflamed tissues,25 and bactericidal capacity.27 Recently, Toll-like receptor–dependent and MyD88-dependent signals from the microbiota were found to drive the phenotypic aging of circulating neutrophils, corresponding with enhanced inflammatory capacity and trafficking from the bloodstream.11 This influence on aging did not correspond with effects on peripheral neutrophil clearance and was independent of Nod1 signaling. Our work affirms that Nod1 plays no role in the intravascular aging of neutrophils, but rather governs the rates of their apoptosis and systemic turnover. Collectively, these results suggest that distinct, parallel signaling mechanisms exist through which the microbiota simultaneously regulates neutrophil aging in the circulation and clearance in the periphery to achieve a balanced equilibrium of neutrophil persistence at homeostasis.
We focused on cell turnover because, in addition to regulating cell abundance directly, mounting evidence suggests phagocyte elimination drives regulatory circuits that feed back on other components of myeloid homeostasis in vivo, including neutrophil development in the bone marrow. Phagocytosis of apoptotic neutrophils by macrophages has been shown to suppress IL-23 production in these cells, thereby inhibiting IL-17A liberation by tissue-resident lymphocytes and suppressing granulopoiesis and neutrophilia.59 More recent studies have shown that rhythmic migrations of senescent neutrophils to the bone marrow modulate the hematopoietic niche to more broadly disfavor progenitor cell retention and maturation.10 Therefore, although alterations of the microbiota can disrupt multiple aspects of myeloid cell biology to impair phagocyte responses, whether these are primary phenomena or secondary to enhanced turnover remains an area of active investigation.
In addition to neutrophils, the stimulatory effect of the flora on cell longevity extended to Ly6C+ IMs, the second-most abundant and short-lived circulating phagocyte.29 The potential fates of IMs are more diverse than that of neutrophils. Although the microbiota has been shown to stimulate monocyte and macrophage development,23 considerably less is known about the mechanisms guiding their turnover. Ly6C+ IMs have been shown to differentiate into long-lived Ly6C−CX3CR1+ monocytes, which patrol vascular endothelia, and into resident macrophages and dendritic cells populating some tissues.29,60,61 However, in addition to inhabiting the first stage of a developmental sequence of mononuclear phagocytes, IMs can emerge from the bone marrow and turn over in tissues at homeostasis while maintaining monocyte identity.62 In contrast, tissue-resident macrophages and dendritic cells arise from a number of cellular origins, including nonhematopoietic cells seeded during embryonic development.29 We focused our investigations on circulating PMNs and IMs because their short lifespans contrast with the more persistent nature of tissue-resident phagocytes and were therefore more likely to be regulated dynamically by tonic microbial signals.
Our finding that neomycin recapitulates the impact of broad-spectrum antibiotics on phagocyte lifespan has precedents in other investigations on the immunomodulatory effects of the microbiota, namely in regulating antiviral responses to influenza infection in the lungs.63 Despite its clinical utility,64 the precise nature and kinetics of neomycin-induced shifts in commensal community structure have not been explored previously. We found that neomycin treatment suppressed the abundance of lactobacilli in the murine intestine, a family of microbes previously shown to bear Nod1 ligands and activate Nod1 in vitro.47,48 This decrement was matched with a compensatory increase in the abundance of Bacteroidetes species, many of which also generate Nod1 ligands. However, detailed analyses of the radial distribution of microbes in the gut suggest that obligate anaerobic bacteria, including Bacteroidetes species, reside predominantly in the luminal center of the intestinal tract.65 Lactobacilli and other oxygen-tolerant bacteria often reside in closer contact with the mucosal surfaces, where oxygen tension is greater and microbial products are sensed more readily by the epithelium.66 This redistribution may explain the decrement in Nod1 signaling observed upon treatment with neomycin.
We determined that stimulation of Nod1 signaling enhanced the abundance of intestinal lymphocytes producing IL-17A and that liberation of this proinflammatory cytokine was required to relay microbial detection at the intestine into systemic changes in phagocyte lifespan. We found γδ T cells to be particularly responsive to Nod1 stimulation, consistent with previous work showing neomycin-sensitive microbiota driving expansion of these IL-17A+ cells.67 IL-17A has been shown to delay the apoptosis of neutrophils and mononuclear phagocytes largely through stimulation of downstream cytokines, including granulocyte colony-stimulating factor and granulocyte-macrophage colony-stimulating factor.54,68 Because the signaling pathways downstream of IL-17A diverge by target cell type and may have overlapping functions, we chose to focus on IL-17A as the most proximal cytokine driving phagocyte survival and therefore the most likely to be amenable to therapeutic intervention.
Advancing scientific understanding of host-commensal relationships has led to intense interest in manipulating the flora to obtain healthful microbial communities. Promising interventions include transplantation of exogenous microbial communities into the gut environment, which has been shown to improve disease outcomes in some severe intestinal infections.69 Our findings imply these interventions may improve the persistence and function of phagocytes that monitor foreign threats throughout the body, providing systemic benefits from local manipulations of microbial communities. Furthermore, growing understanding of the central importance of apoptosis and cell turnover to immune homeostasis has spurred an increase in the development of therapeutics designed to target this component of phagocyte physiology directly. For example, inhibitors of cyclin-dependent kinases have been shown to improve resolution of pathogenic pulmonary inflammation by hastening the apoptosis of infiltrating neutrophils.70 Our results suggest that the ecology of the intestinal microbiota may modulate the efficacy of this therapy and others like it, potentially providing an opportunity for adjuvantive therapies targeting specific gut microbial taxa to improve the outcome of patients with diverse inflammatory diseases.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Kristin Blouch and Susan Ross for Tlr4−/− mice; Mark Boyer and Sunny Shin for mice and reagents; and Jamie Knox, Michael Betts, Laura Vella, and E. John Wherry for flow cytometry reagents.
This work was supported by grants from the National Institute of Allergy and Infectious Diseases, National Institutes of Health (grants AI038446, AI105168, and AI060516 [J.N.W.]; AI045008 [F.D.B.]); Fundacao de Amparo a Pesquisa do Estado de Sao Paolo (FAPESP) (award 2014/00027-9) (E.T.); and National Heart, Lung, and Blood Institute, National Institutes of Health (grant HL113252) (F.D.B.). T.B.C. is a Sir Henry Dale Fellow jointly funded by the Wellcome Trust and Royal Society (grant 107660/Z/15/Z). The authors also acknowledge support from the PennCHOP Microbiome Program.
Authorship
Contribution: C.B.H. and J.N.W. designed the research; C.B.H. performed the majority of the experiments, with E.T. and A.M.R. contributing to collection and analysis of microbial samples; T.B.C. provided essential reagents; A.L., A.G.B., and F.D.B. performed and analyzed metagenomic sequencing experiments; C.B.H., F.D.B., and J.N.W. analyzed and interpreted experimental findings; C.B.H. and J.N.W. wrote the paper with important contributions from F.D.B.; and all authors reviewed the manuscript before submission.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jeffrey N. Weiser, Medical Science Building, Room 256, 550 First Ave, New York, NY 10016; e-mail: jeffrey.weiser@nyumc.org.