Abstract
Response assessment in lymphoma relies on imaging scans that do not capture biologic processes at the molecular level. Monitoring circulating tumor DNA (ctDNA) with next-generation sequencing–based assays can detect recurrent disease prior to scans and “liquid biopsies” for somatic mutations address tumor heterogeneity, clonal evolution, and mechanisms of resistance to guide precision treatment. Preanalytic collection and processing procedures should be validated and standardized. We describe emerging applications of ctDNA monitoring including real-time analysis of tumor dynamics, preclinical disease detection, and precision-directed treatment paradigms.
Introduction
Monitoring treatment response in non-Hodgkin lymphoma relies on computerized tomography (CT) and 18fluoro-2-deoxyglucose positron emission tomography (PET) scans. Imaging scans provide macro estimates of tumor volume and location, but do not significantly improve survival and are limited by cost and risk of ionizing radiation.1-3 Because disease recurrence originates from persistent tumor below limits of clinical detection, imaging scans are suboptimal for surveillance monitoring in curable lymphomas, such as diffuse large B-cell lymphoma (DLBCL).1 Lack of specificity limits the accuracy of PET scans for response-adapted approaches and surveillance, despite improved sensitivity over CT scans.4,5 Imaging scans are “snapshots” of clinically detectable tumor and cannot capture dynamic processes such as tumor response kinetics, clonal evolution, and cellular resistance.
With emergence of targeted therapy, clinically validated technology is needed to temporally assess the molecular heterogeneity across disease sites.6,7 Tissue biopsies determine molecular features of a tumor, but are prone to sampling bias and difficult to obtain serially. Circulating tumor DNA (ctDNA) can provide an average of the overall clonal heterogeneity and noninvasively assess molecular changes over time. Free from sampling bias of singular site biopsies, ctDNA integrates all genetic lesions and provides a more detailed view of the tumor.
Assessment of ctDNA is powerful technology that overcomes fundamental limitations of imaging scans and molecular analysis via tissue biopsies. In this review, we discuss the technical issues and present the promises of ctDNA for clinical application and novel research design.
Technical aspects of ctDNA
Fragmented DNA from normal and diseased tissue is constantly shed into the bloodstream as cell-free DNA (cfDNA) through processes of apoptosis, necrosis, and secretion.8,9 Patients with cancer have higher overall levels of cfDNA than healthy people, but the fraction of DNA from malignant cells, captured as ctDNA, may be as low as 0.01%.10-12 Because ctDNA derives from tumor tissue, it is a highly specific tumor biomarker.13 It is present without detectable circulating tumor cells and correlates with tumor burden in early- and late-stage malignancies.14,15 Assays designed for molecular monitoring must accurately discriminate ctDNA from DNA that originates in nonmalignant tissue.16
Molecular monitoring of lymphoma by ctDNA has gained momentum as a result of technical advances in detection capability and processing speeds (Table 1). Quantification of ctDNA in B-cell lymphomas using polymerase chain reaction (PCR) analysis of rearranged immunoglobulin heavy chains are subject to relatively low sensitivity and artifact, limiting their clinical utility.17 Modern platforms combine universal PCR primers for the VDJ regions of immunoglobulin receptors with next-generation sequencing (NGS), resulting in highly sensitive and specific detection of ctDNA in multiple B-cell lymphomas.15,18,19 As a surrogate for the entire tumor genome, ctDNA may also be analyzed for tumor-specific mutations, commonly referred to as a “liquid biopsy.”20,21 Modern NGS techniques have the necessary specificity, and bandwidth to identify genetic aberrations circulating at low allele frequencies.22,23
Methods for molecular monitoring of ctDNA for non-Hodgkin lymphoma
| . | ctDNA of IgH . | ctDNA of VDJ sequence . | Tumor genotypic ctDNA . |
|---|---|---|---|
| Technique | Allele-specific PCR | PCR + NGS | NGS |
| Primary tumor required | Yes | No* | No† |
| Sensitivity‡ | 1 in 105 | 1 in 106 | Unknown |
| Processing time | Weeks | 1-2 wk | 1-2 wk |
| Track clonal evolution | No | Limited | Possible |
| Track resistance | No | Limited | Possible |
| Potential | Tumor-specific Quantifiable | Tumor-specific Quantifiable Universal primers Rapid turnaround time | Broad genomic coverage Track clonal evolution Track resistance mechanisms |
| Limitations | Not universal Specific primers required Limited foci assessed | Limited genotypic information | Low allele frequency Molecular heterogeneity of tumor |
| . | ctDNA of IgH . | ctDNA of VDJ sequence . | Tumor genotypic ctDNA . |
|---|---|---|---|
| Technique | Allele-specific PCR | PCR + NGS | NGS |
| Primary tumor required | Yes | No* | No† |
| Sensitivity‡ | 1 in 105 | 1 in 106 | Unknown |
| Processing time | Weeks | 1-2 wk | 1-2 wk |
| Track clonal evolution | No | Limited | Possible |
| Track resistance | No | Limited | Possible |
| Potential | Tumor-specific Quantifiable | Tumor-specific Quantifiable Universal primers Rapid turnaround time | Broad genomic coverage Track clonal evolution Track resistance mechanisms |
| Limitations | Not universal Specific primers required Limited foci assessed | Limited genotypic information | Low allele frequency Molecular heterogeneity of tumor |
IgH, immunoglobulin heavy chain; NGS, next-generation sequencing; PCR, polymerase chain reaction; VDJ, variable-diversity-joining region of the immunoglobulin receptor.
Tumor clonotype can be determined without baseline tissue, but the yield is lower.
Mutational panels common to lymphoma subtypes would obviate the need for baseline tumor.
Sample size required to detect 1 cellular equivalent.
Methods to identify specific ctDNA sequences perform best on tumor biopsies. Clonal VDJ sequences can be determined from baseline tumor tissue in more than 85% of DLBCL cases, but with a significantly lower yield from blood samples.15,19,24 Targeted resequencing mutational panels for ctDNA are now emerging, but also require baseline biopsy samples to confirm tumor origin.22,23,25 Hence, quality DNA extraction from pretreatment tissue is important and is affected by low tumor content and necrosis.
Within peripheral blood samples, high rates of DNA fragmentation and low ratios of tumor DNA are methodologically challenging barriers. Normal DNA contamination from white blood cells is typically higher in the serum than plasma, and delay and temperature of blood samples before centrifugation affect DNA concentration.26 Specialized collection tubes (Streck) can reduce DNA degradation by nucleases and contamination from white blood cell DNA.27 Although successful analysis of ctDNA can be performed on stored serum samples, standardized collection procedures will optimize the analysis, particularly for detection of tumor mutations.26,28
Monitoring tumor genotype
The complete molecular heterogeneity of a tumor cannot be adequately assessed by single or even multiple biopsies, whereas a “liquid biopsy” captures genetic information shed from all sites of disease.16 Recent reports demonstrate high concordance between mutations within tissue and targeted resequencing panels from ctDNA23,25 Analysis of ctDNA can also detect somatic mutations not identified in tumor biopsies. However, mutations with low allele frequencies are difficult to detect in the blood, suggesting that the different methods provide complementary information. Liquid biopsies of ctDNA can also be effective when needle biopsies are challenging, such as isolated central nervous system disease.15,19,29 Recent reports have demonstrated that tumor-associated mutations are detectable in the plasma and cerebral spinal fluid of patients with primary central nervous system lymphoma (including L.M.S. and W.H.W., unreported data).30
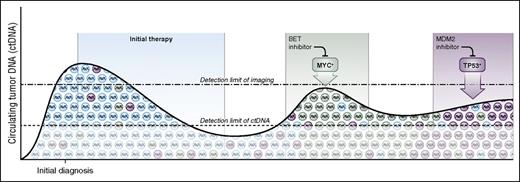
In addition to spatial heterogeneity, lymphomas exhibit temporal heterogeneity and continuously evolve over time, particularly under treatment selection pressure (Figure 1). In the case of DLBCL, evidence suggests recurrent disease comprises multiple genetically distinct subclones. Serial analysis of ctDNA for mutation allele frequency can analyze clonal evolution in consecutive samples and reveal a shift in the dominant subclone with potential implications for treatment.31-33 Monitoring ctDNA is a promising method for assessing subclone allele frequency to overcome both spatial and temporal tumor heterogeneity.7
Serial monitoring with circulating tumor DNA to guide precision medicine. Lymphomas are composed of multiple tumor clones and subclones that serially evolve over time, especially under the selective pressure of therapy. Liquid biopsies of ctDNA detect molecular features of resistant disease at the molecular level and can noninvasively genotype ctDNA throughout the disease course. Serial monitoring of ctDNA may be a powerful tool to address temporal heterogeneity of tumors, to detect clonal evolution, and to study mechanisms of treatment resistance. The timing and nature the dominant relapsing clone may guide precision treatment at relapse. As examples, patients who relapse with a myc rearrangement (green) might be offered targeted therapy with a BET inhibitor, whereas patients who relapse with a TP53 mutation (magenta) could be offered an MDM2 inhibitor.
Serial monitoring with circulating tumor DNA to guide precision medicine. Lymphomas are composed of multiple tumor clones and subclones that serially evolve over time, especially under the selective pressure of therapy. Liquid biopsies of ctDNA detect molecular features of resistant disease at the molecular level and can noninvasively genotype ctDNA throughout the disease course. Serial monitoring of ctDNA may be a powerful tool to address temporal heterogeneity of tumors, to detect clonal evolution, and to study mechanisms of treatment resistance. The timing and nature the dominant relapsing clone may guide precision treatment at relapse. As examples, patients who relapse with a myc rearrangement (green) might be offered targeted therapy with a BET inhibitor, whereas patients who relapse with a TP53 mutation (magenta) could be offered an MDM2 inhibitor.
Monitoring tumor VDJ sequence
DLBCL is molecularly heterogeneous, but rearranged VDJ immunoglobulin receptor genes are unique to each patient’s tumor and are readily detectable by modern NGS platforms.34 Quantitative high-throughput methods that combine PCR-based amplification of immunoglobulin gene segments and NGS can detect and quantify ctDNA from B-cell lymphomas in the blood with a detection limit of 1 tumor cell equivalent per 106 diploid genomes.15,19
A recent study in 126 patients with untreated DLBCL demonstrated that ctDNA of VDJ predicted early treatment failure and serial monitoring after therapy identified disease recurrence months before CT scans.15 Another study in a separate cohort of patients with DLBCL demonstrated that monitoring cell-free ctDNA of VDJ in plasma was more effective than monitoring circulating cells with the same assay.19 Notably, both studies showed a significant association between the quantity of pretreatment ctDNA and indices of tumor mass.
Applications of ctDNA monitoring
Clinical applications of ctDNA depend on the assay performed, the timing of the assay, and the goal of therapy (Table 2). The therapeutic goal for aggressive B-cell lymphomas, such as DLBCL, is cure. This requires eradication of all tumor clones, or disease recurrence is virtually inevitable. Molecular relapse of ctDNA heralds failure of curative treatment. The highly sensitive and quantitative characteristics of VDJ ctDNA lead to early detection of treatment failure compared with conventional imaging (Figure 2A).15,19 Indeed, identification of a molecular relapse allows institution of salvage treatment before the development of high disease burden that is required for clinical detection of recurrence.
Clinical applications of circulating tumor DNA for non-Hodgkin lymphoma
| . | Pretreatment . | Receiving treatment . | End-of-treatment . | After treatment . |
|---|---|---|---|---|
| ctDNA of VDJ sequence | Prognostic tool | Quantitate tumor kinetics Early treatment failure Response-adapted therapy | Molecular remission Maintenance therapy decisions | Early recurrence detection Clonal evolution |
| Tumor genotypic ctDNA | Prognostic tool | Quantitate tumor kinetics | Molecular remission | Early recurrence detection |
| Initial therapy choice | Early treatment failure Early resistance mechanisms Response-adapted therapy | Maintenance therapy decisions | Clonal evolution Late resistance mechanisms Select next therapy |
| . | Pretreatment . | Receiving treatment . | End-of-treatment . | After treatment . |
|---|---|---|---|---|
| ctDNA of VDJ sequence | Prognostic tool | Quantitate tumor kinetics Early treatment failure Response-adapted therapy | Molecular remission Maintenance therapy decisions | Early recurrence detection Clonal evolution |
| Tumor genotypic ctDNA | Prognostic tool | Quantitate tumor kinetics | Molecular remission | Early recurrence detection |
| Initial therapy choice | Early treatment failure Early resistance mechanisms Response-adapted therapy | Maintenance therapy decisions | Clonal evolution Late resistance mechanisms Select next therapy |
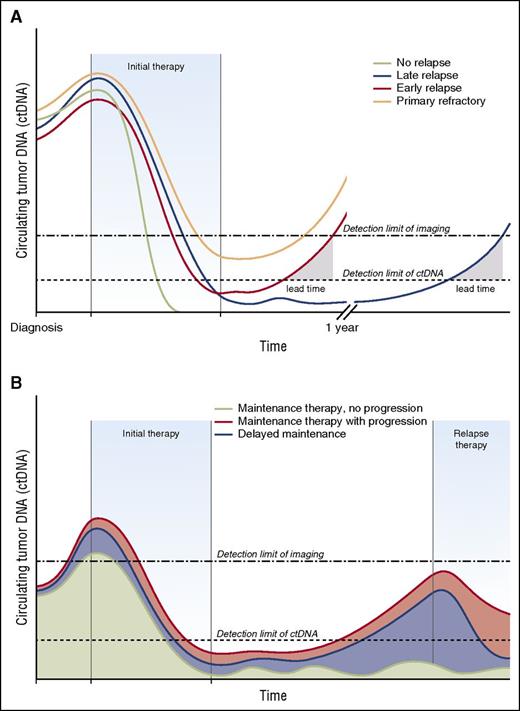
Monitoring circulating tumor DNA enhances detection of relapse and defines molecular remission. The lead time offered by serial monitoring of ctDNA represents an opportunity for early intervention with minimal tumor burden. The clinical applications would differ when treating with curative intent (ie, aggressive lymphomas) or more extended duration of therapy (ie, indolent lymphomas). (A) Monitoring therapy for ctDNA for curative intent. The patient with no relapse (green) achieves a complete molecular remission and represents successful cure of lymphoma. The patient with late relapse (blue) initially achieves a complete molecular remission, but has ctDNA reappear before imaging, which creates a lead time for possible intervention. The patient with early relapse (red) has rising levels of ctDNA shortly after completion of therapy with a narrower lead time. The patient with primary refractory disease (brown) has persistence of minimal residual disease at the end of therapy that is undetectable by imaging. (B) Monitoring therapy with ctDNA for extended duration. Indolent lymphomas are frequently treated for extended durations with maintenance therapy designed to prolong duration of remission. Successful maintenance therapy (green) could be monitored with ctDNA and continued as long as disease remains undetectable. Patients who have the reappearance of ctDNA while receiving maintenance therapy (red) might be considered for alternative therapy before clinical effects. Patients who are not initially treated with maintenance therapy can be offered “delayed maintenance” at a time when disease is detectable by ctDNA, but not yet detectable by imaging scans.
Monitoring circulating tumor DNA enhances detection of relapse and defines molecular remission. The lead time offered by serial monitoring of ctDNA represents an opportunity for early intervention with minimal tumor burden. The clinical applications would differ when treating with curative intent (ie, aggressive lymphomas) or more extended duration of therapy (ie, indolent lymphomas). (A) Monitoring therapy for ctDNA for curative intent. The patient with no relapse (green) achieves a complete molecular remission and represents successful cure of lymphoma. The patient with late relapse (blue) initially achieves a complete molecular remission, but has ctDNA reappear before imaging, which creates a lead time for possible intervention. The patient with early relapse (red) has rising levels of ctDNA shortly after completion of therapy with a narrower lead time. The patient with primary refractory disease (brown) has persistence of minimal residual disease at the end of therapy that is undetectable by imaging. (B) Monitoring therapy with ctDNA for extended duration. Indolent lymphomas are frequently treated for extended durations with maintenance therapy designed to prolong duration of remission. Successful maintenance therapy (green) could be monitored with ctDNA and continued as long as disease remains undetectable. Patients who have the reappearance of ctDNA while receiving maintenance therapy (red) might be considered for alternative therapy before clinical effects. Patients who are not initially treated with maintenance therapy can be offered “delayed maintenance” at a time when disease is detectable by ctDNA, but not yet detectable by imaging scans.
Indolent lymphomas present a different scenario because they generally are not treated with curative intent. In this setting, monitoring of ctDNA provides a quantitative estimate of sensitivity to treatment and can be monitored serially to guide treatment decisions. The common practice to treat indolent lymphomas for prolonged and even indefinite periods of time offers opportunities for ctDNA monitoring (Figure 2B). In some cases, extended treatment may be unnecessary, such as in patients who become ctDNA negative (ie, minimal residual disease negative), whereas in other cases, resistant clones may accumulate below the level of imaging detection. In both cases, serial monitoring of ctDNA could inform clinical decision-making, including treatment duration, retreatment timing, and guide precision treatment based on features of the dominant clones (Figure 1).
Pretreatment applications
ctDNA resequencing mutational panels (liquid biopsies) and quantification of pretreatment VDJ ctDNA provide novel opportunities for precision treatment. Multiple studies have shown a correlation between clinical determinants of tumor burden and quantitative ctDNA.14,15,19 A few studies have reported that higher pretreatment concentrations of ctDNA are associated with a poorer prognosis, possibly because of higher tumor burden, and hence the potential for more aggressive treatment.11,23 Molecular features such as tumor metabolism and/or proliferation may also be captured by ctDNA analysis and provide new areas of investigation. The molecular information obtained from liquid biopsies may provide valuable information on targetable oncogenic pathways and mutations, which are heterogeneous in DLBCL and often track with the cell of origin.6
Response assessment
The current response criteria for lymphoma rely on CT and PET imaging, and the completeness of response to initial therapy is prognostic.35,36 Detection of subclinical disease by ctDNA, however, could identify patients not in complete molecular remission (Figure 2A). A recent study of ctDNA for tumor VDJ in DLBCL showed that patients who did not achieve a complete molecular remission were not cured, indicating its importance in aggressive lymphomas.15 For indolent lymphomas, achieving complete molecular remission may assist with treatment decisions regarding maintenance therapy and provide a novel endpoint for clinical trials and drug approval. In follicular lymphoma, PCR-based primers to detect BCL2 breakpoints have been used to assess molecular remission, which is associated with improved outcome.37,38 Similar findings have been reported in mantle cell lymphoma, using PCR assays to detect the CCND1/IgH translocations.39 These findings support the importance of molecular remissions.
Interim monitoring
Interim ctDNA monitoring (ie, during treatment), such as interim PET scans, can be developed for risk-adapted treatment strategies including early treatment termination for patients in molecular remission or treatment modification for patients with poorly responsive tumors. Although risk-adaptive strategies based on interim PET scans can identify patients with DLBCL at risk for treatment failure, 2 recent prospective studies failed to show clinical benefit from switching therapy, indicating the need for more sensitive and specific selection methods.40-42 Interim ctDNA monitoring has potential advantages over PET scans as a result of high tumor specificity, quantitative analysis, and temporal kinetics. Indeed, quantitative response kinetics may improve on response-adapted strategies in lymphomas. Our group studied the predictive significance of quantitative ctDNA for VDJ after each treatment cycle in previously untreated patients with DLBCL.15 Patients who cleared ctDNA after 2 cycles were significantly more likely to be progression-free at 5 years compared with patients who were ctDNA positive (80.2% vs 41.7%; P < .0001).15 Approximately half the patients cleared ctDNA after only 1 cycle, and 78% were negative after 2 treatment cycles. Various patterns of interim ctDNA kinetics were associated with early treatment failure, but absence of clearance was associated with the shortest survival.15 Other reports of interim ctDNA monitoring in DLBCL have also observed that ctDNA concentrations after only 1 to 2 cycles are predictive.22 Future studies are needed to validate the clinical use of interim ctDNA and its role with interim PET scans.
Posttreatment monitoring
The ability to identify patients with aggressive lymphomas before they develop clinically detectable disease has significant appeal because some may yet be cured with further treatment (Figure 2A). Although this has not been prospectively validated, it has been shown that patients with lower disease burdens at recurrence have better outcomes.43 The potential value of surveillance ctDNA monitoring lies in its low detection limit and ease of repetition. We showed that serial surveillance monitoring of ctDNA for VDJ after therapy in DLBCL detected recurrence a median of 3.5 month (range, 0-200 months) before CT scans.15 In patients who relapsed more than 6 months from the end of therapy, ctDNA was identified in 91% (10/11), and 80% (8/10) were detected before clinical disease on CT scans. In another series, similar rates of detection before relapse were observed.19 Paradigms for molecular surveillance monitoring will differ according to lymphoma subtype and therapeutic goals. For indolent lymphoma, the usual clinical goal is disease control, although the potential for cure cannot be ruled out. In the case of long-term disease control, despite the persistence of malignant clones, the goal of surveillance monitoring is to assess the malignant clone kinetics clonal evolution (Figure 1).
ctDNA and precision medicine
Identification of the molecular aberrations within an individual patient’s lymphoma will be needed for precision treatment decisions.6 Barriers to precision treatment include spatial and temporal tumor heterogeneity.7 Although in its infancy and unvalidated for clinical decision-making, the liquid biopsy has obvious advantages for selecting treatment based on the identification of dominant resistant clones (Figure 1). For example, increasing frequency of clones with myc translocations or TP53 mutations suggest emerging resistance, and may allow treatment modifications with targeted agents before the emergence of clinically detectable disease (Figure 1). Individual somatic mutations can also predict response to target agents, as recently shown with CD79B and MYD88 mutations in DLBCL and response to ibrutinib.44 Such information is likely to significantly improve outcomes through rational timing of treatment and precision drug selection. A new generation of “smart trials” based on this technology may also lead to more rational and rapid drug development and approval.
Conclusions
Circulating tumor DNA has the potential to transform clinical care paradigms and future trial designs. The ability of tumor VDJ ctDNA to dynamically assess molecular tumor response and detect recurrence of occult disease in B-cell lymphomas will undoubtedly improve the care of these patients. For indolent lymphomas, ctDNA provides a critical “look below the water line” that can aid clinical decisions on treatment timing and duration. Liquid biopsies of tumor genotypic ctDNA add a further dimension that can integrate all the genetic lesions within a tumor and optimally address traditional barriers to precision treatment such as tumor heterogeneity and clonal evolution. As we move forward, it will be imperative to standardize collection, storage, and processing procedures for ctDNA technology and validate its clinical and research utility.
Acknowledgments
We acknowledge support from the intramural research program of the National Institutes of Health, especially Kieron Dunleavy and the research team. Alan Hoofring and S. Peter Wu are acknowledged for illustration support.
Authorship
Contribution: All authors (M.R., L.M.S., and W.H.W.) made a substantial contribution to discussion of the content. M.R. wrote the first draft of the manuscript, and W.H.W. revised the final draft. All authors reviewed and edited the final manuscript before submission.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mark Roschewski, Lymphoid Malignancies Branch, Center for Cancer Research, National Cancer Institute, National Institutes of Health, 9000 Rockville Pike, Bldg 10, Room 4N/115, Bethesda, MD 20892; e-mail mark.roschewski@nih.gov.