Key Points
Immunoassays used to diagnose heparin-induced thrombocytopenia vary substantially with regard to the specific test characteristics.
High sensitivity (>95%) in combination with high specificity (>90%) was found in only 5 tests.
Abstract
Immunoassays are essential in the workup of patients with suspected heparin-induced thrombocytopenia. However, the diagnostic accuracy is uncertain with regard to different classes of assays, antibody specificities, thresholds, test variations, and manufacturers. We aimed to assess diagnostic accuracy measures of available immunoassays and to explore sources of heterogeneity. We performed comprehensive literature searches and applied strict inclusion criteria. Finally, 49 publications comprising 128 test evaluations in 15 199 patients were included in the analysis. Methodological quality according to the revised tool for quality assessment of diagnostic accuracy studies was moderate. Diagnostic accuracy measures were calculated with the unified model (comprising a bivariate random-effects model and a hierarchical summary receiver operating characteristics model). Important differences were observed between classes of immunoassays, type of antibody specificity, thresholds, application of confirmation step, and manufacturers. Combination of high sensitivity (>95%) and high specificity (>90%) was found in 5 tests only: polyspecific enzyme-linked immunosorbent assay (ELISA) with intermediate threshold (Genetic Testing Institute, Asserachrom), particle gel immunoassay, lateral flow immunoassay, polyspecific chemiluminescent immunoassay (CLIA) with a high threshold, and immunoglobulin G (IgG)-specific CLIA with low threshold. Borderline results (sensitivity, 99.6%; specificity, 89.9%) were observed for IgG-specific Genetic Testing Institute-ELISA with low threshold. Diagnostic accuracy appears to be inadequate in tests with high thresholds (ELISA; IgG-specific CLIA), combination of IgG specificity and intermediate thresholds (ELISA, CLIA), high-dose heparin confirmation step (ELISA), and particle immunofiltration assay. When making treatment decisions, clinicians should be a aware of diagnostic characteristics of the tests used and it is recommended they estimate posttest probabilities according to likelihood ratios as well as pretest probabilities using clinical scoring tools.
Introduction
In clinical practice, immunoassays are pivotal for the workup of patients with suspected heparin-induced thrombocytopenia (HIT).1-4 HIT is a life-threatening complication of heparin therapy that affects a significant number of patients.5 It is associated with a high morbidity and mortality because of a massive pro-coagulant state, with a high incidence of extensive venous and arterial thrombosis, limb loss, and even death.6,7 Suspicion of HIT requires an immediate diagnostic workup to prevent severe complications.1,8,9 Still, diagnosis of HIT is challenging.1,10 Functional assays, such as the serotonin release assay (SRA) or the heparin-induced platelet activation assay (HIPA) are accepted as gold standard, but are rarely available in a timely manner.2,3,11 Clinical assessment tools such as the thrombocytopenia, thrombosis, timing of decrease in platelet count, and other causes for thrombocytopenia (4Ts) score can exclude HIT in many patients if conducted by experienced observers.12 However, the positive predictive value is low,12 they are subject to a relevant inter-observer variability,13 and they are not adequately evaluated in all settings.14 To make a treatment decision at bedside, physicians are recommended by current guidelines and recent reviews to order an HIT immunoassay in all patients with an intermediate or high-risk 4Ts score.1,8,10-12,15
Many tests and several classes of assays have been developed by manufacturers. They recognize different classes of antibodies, use varying thresholds, and may apply test variations such as the high-dose heparin confirmation step. All tests are based on detecting antibodies targeting complexes of platelet factor 4 (PF4) bound to heparin (or other polyanions respectively).16,17 A solid-phase enzyme-linked immunosorbent assay (ELISA) using a low optical-density threshold (OD) and targeting immunoglobulin G (IgG), IgM, and IgA antibodies was developed in the mid-1990s.16,18 Later, other tests were introduced: IgG-specific assays,19 high- and intermediate-OD thresholds,19,20 and the high-dose heparin confirmation step.21 Other classes of assays have been developed to enable short turnaround times and a 24-hour service: particle gel immunoassay (PaGIA), particle immunofiltration assay (PIFA), lateral flow immunoassay, latex agglutination assay, and chemiluminescent immunoassay (CLIA). Different antibody specificities are available for CLIA, and several thresholds have been proposed for CLIA and PaGIA. The corresponding diagnostic accuracy studies are hard to interpret because of varying results, imprecise estimates, and sometimes conflicting data. In clinical practice, it is difficult to choose the right assay because it remains unclear if and how test characteristics influence the diagnostic value. Several authors have raised concerns that the diagnostic value may differ relevantly among the individual tests.2,22-24 As a result of this uncertainty, the application of assays varies greatly among laboratories.25
The aim of the present investigation was to assess the diagnostic accuracy with regard to different classes of assays, antibody specificities, thresholds, manufacturers, application of a high-dose heparin confirmation step, and material used. In addition, we assessed whether different reference standards used in diagnostic accuracy studies have an impact on diagnostic accuracy. We retrieved all available data, applied strict inclusion criteria, systematically assessed methodological quality, and pooled diagnostic accuracy measures if possible.
Methods
Before starting the investigation, we developed a research protocol according to recommendations of the Cochrane Diagnostic Test Accuracy Working Group.26
Study identification
A search strategy was developed for the Medline, EMBASE, and Cochrane Collaboration databases to identify diagnostic accuracy studies of immunoassays for diagnosis of HIT (see supplemental data on the Blood Web site). Search strategy was refined using keywords of references found in a pilot search and after manual review of reference lists. Search strategy was tested in 12 index publications (100% sensitivity). The literature search was supplemented by manual review of reference lists, including the American College of Chest Physicians Evidence-Based Clinical Guidelines (9th edition).2 No restrictions or filters with regard to language, publication date, or age range were applied. The last search run was conducted on November 2, 2014. Records were screened by 2 investigators (M.N., A.t.C.-H.); all hits were assessed in full text for eligibility.
Study eligibility
The following inclusion criteria were applied: (1) evaluation of a commercially available or fully described immunoassay to detect heparin/PF4 antibodies; (2) application of the index test to patients with suspected HIT; (3) application of a reference standard that is established or fully described; and (4) numbers of true positives, false positives, true negatives, and false negatives are reported or can be calculated. We considered a reference standard test to be established if it was evaluated in reasonably designed studies and implemented in routine practice. Studies were excluded in case of (1) application of the index test to patients without any suspicion for HIT (screening test), (2) evaluation of tests that are not immunoassays, (3) evaluation of confirmatory tests after application of a previous laboratory test, (4) reference standard not clearly defined, and (5) studies that compare diagnostic accuracy of 1 immunoassay with another 1 without using an established reference standard. Two reviewers assessed eligibility and consensus was achieved by discussion (M.N., A.t.C.-H.; raw agreement, 0.91; Cohen’s κ, 0.71).
Data extraction
Included studies were reviewed in duplicate and the following data were extracted: author; year of publication; study design; type of patient selection; characteristics of study population including cohort criteria, inclusion procedure, and setting; characteristics of index test including antibody specificity, thresholds, material used, and use of high-dose heparin confirmation test; application of index test; characteristics of reference standard; application of reference standard; sample size; disease prevalence; and numbers of true positives, false positives, true negatives, and false negatives (reported or calculated). Test results were categorized according to the “intention-to-diagnose” principle to avoid biased overestimation of diagnostic accuracy27 : inconclusive results of the index test were classified as negative if the reference standard was positive and were rated as positive if the reference standard was negative.27 Observations were excluded from analysis if the reference standard revealed inconclusive results. For example, in 1 study, an inconclusive PIFA result was classified as negative because SRA was positive, another inconclusive PIFA result was counted as positive because SRA was negative, and 2 observations were excluded because of a borderline SRA result in combination with a negative ELISA and a low 4T score.28 Two-by-two tables were created for every evaluation study. Sensitivity, specificity, positive likelihood ratio, negative likelihood ratio, and pretest probability (prevalence) were calculated. To account for differences in the scale of the threshold among different assays (ELISA assays in particular), we categorized thresholds into low, intermediate, and high. For ELISA, ODs ≤0.7 were classified as low, OD between 0.8 and 1.4 as intermediate, and OD >1.4 as high. A positive/negative PaGIA result was ranked as low, and a titer of 2 or 3 as intermediate. LFI results were categorized as low (no studies using titration available). For CLIA, a threshold of 1.0 U/mL was classified as low, between 1.0 and 2.8 U/mL as intermediate, and above 2.8 U/mL as high. Threshold of latex agglutination assay (1.0 U/mL) was classified as low.
Assessment of methodological quality
The methodological quality of the individual studies was assessed in terms of risk of bias and concerns regarding applicability using the revised tool for quality assessment of diagnostic accuracy studies (QUADAS-2). Details of this validated and widely accepted instrument are published elsewhere.29 Then, methodological quality was assessed in 4 domains (patient selection, index test, reference standard, flow and timing) by using signaling questions. According to the published guidelines, we adjusted the signaling questions to our particular research questions and developed detailed decision criteria. In a pilot study, the adapted QUADAS-2 tool was applied to a subset of 7 studies by 2 investigators (M.N., A.t.C.-H.) with good agreement. Signaling questions and decision criteria were refined after these pilot studies to further improve reliability. Application of adapted QUADAS-2 tool to all included studies was done in duplicate and disagreement was solved by discussion between 2 investigators.
Statistical analysis
To assess the diagnostic accuracy of different classes of assays, we grouped observations by class, antibody specificity, threshold, and the use of the high-dose heparin confirmation step. In addition, we considered differences in ELISA assay designs by grouping according to manufacturer, antibody specificity, threshold, and use of the high-dose heparin confirmation step. To observe the impact of different reference standards used in evaluation studies, we conducted a stratified analysis in the assay class with the most accessible studies and performed a meta-regression analysis (see the following section). To maintain independency of observations, studies that were conducted in the same study population were included only once in each group. In these cases, we selected the study to be included according to the following priorities: (1) the largest number of observations and (2) type of manufacturer that was included less often within the particular category. Because heterogeneity is expected to be high in systematic reviews of diagnostic accuracy studies, we followed current recommendations and did not calculate an I2 statistic.30 Diagnostic accuracy measures were pooled with the use of the unified model,31 comprising a bivariate random-effects model32 and a hierarchical summary receiver operating characteristics model.33 However, it was not possible to fit these models in every category because at least 4 studies are required to perform the analysis. To give an approximation of the diagnostic accuracy in these tests, we fitted a fixed effect model according to Mantel and Haenszel.30
Potential sources of heterogeneity and bias
Several measures were carried out to explore sources of heterogeneity and to recognize potential sources of bias. First, results of studies with a higher methodological quality were compared with estimates obtained from all studies. We selected the criterion of cohort studies (in contrast to case-control studies) because it is considered to have the highest impact on diagnostic accuracy study results.34 Second, we compared measures between tests within the same study population using the same reference standard. Third, estimates obtained with accepted gold standards (SRA/HIPA) were compared with estimates from all studies. Finally, we conducted a random-effects meta-regression35 using diagnostic odds ratio as outcome variables and important characteristics as predictors (type of reference standard [SRA/HIPA vs others], material used [plasma vs serum], setting [surgical vs mixed population; intensive care unit patients vs mixed population]).
All statistical analyses were conducted using Stata 13.1 (StataCorp, 2013, Stata Statistical Software, release 13; College Station, TX), figures of methodological quality were generated using Review Manager (version 5.3, Copenhagen: The Nordic Cochrane Centre, The Cochrane Collaboration, 2014).
Results
Study identification and selection
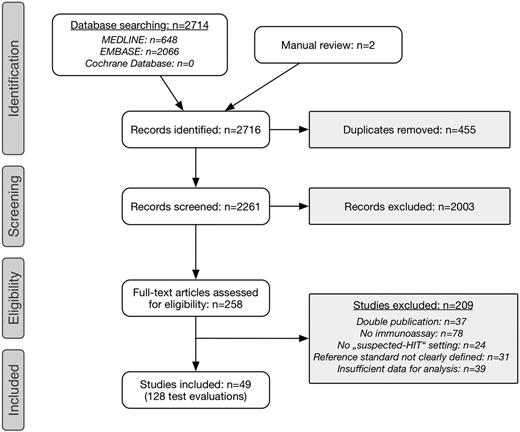
Literature search yielded 2716 records, including 2 publications identified by manual review (Figure 1). After removal of duplicates, title and abstract were screened in 2261 records. We selected 258 publications for full-text review. Of these, 37 were excluded because of duplicate publication (conference abstract as well as original article), 78 studies did not evaluate an immunoassay, 24 investigations were not conducted in patients with suspected HIT, 39 articles because of insufficient data for analysis, and the reference standard was not clearly defined in 31 publications. Finally, we included 49 studies comprising 128 test evaluations in 15 199 patients.16,19-21,28,36-79
Selection process of primary studies. A Preferred Reporting Items for Systematic Reviews and Meta-Analyses flowchart is shown.
Selection process of primary studies. A Preferred Reporting Items for Systematic Reviews and Meta-Analyses flowchart is shown.
Study characteristics
Detailed characteristics of all 128 diagnostic accuracy studies are given in supplemental Table 1. Six different classes of assays were evaluated: ELISA (79 evaluation studies),16,19-21,28,39-41,43-55,59-61,63-79 PaGIA (14),20,36,42,45,46,53,58,64,65,68,70,72,75,79 PIFA (1),28 lateral flow immunoassay (9),54-56,58,66,70,72 latex agglutination assay (1),37 and CLIA (24).37,38,57,61,62,77 Available assays, antibody specificities, thresholds, test variations, and manufacturers are illustrated in Table 1. Often, more than 1 evaluation study was reported in a publication (range, 1-13), and more than 1 assay was evaluated in the same study population (range, 1-11). Low thresholds were studied in 96 cases,16,19-21,28,36-64,66-79 intermediate in 21,19-21,28,47,61,65,77 and high in 11.19,37,39,47,61,62,77 Assays targeting polyspecific antibodies were used in 78 studies,16,19-21,28,36-39,41-43,45-54,57-65,67-75,77-79 and IgG-specific antibodies in 49 investigations19-21,37,40,44,54-58,61,63,64,66,69-72,77 (unclear in 1 study76 ). A high-dose heparin confirmation step (ELISA) was studied in 9 cases.21,40,67,73,76 The following reference standard tests have been used: SRA (43 evaluation studies),19,28,36,38,39,41-43,47,50,53,63,64,67-69,73,74,76 HIPA (42),16,20,21,37,40,48,49,51,52,54-56,70,72 combination of HIPA/SRA with clinical criteria (10),46,57-59,78 flow cytometry (12),44,45,66,77 heparin-induced platelet aggregation test (4),60,65 and others (17).61,62,71,75,79
Immunoassays for diagnosis of HIT: available classes of assays, antibody specificities, thresholds, test variations, and manufacturers
| Classes of assays . | Antibody specificity . | Threshold . | Test variation . | Manufacturer (names of tests) . |
|---|---|---|---|---|
| ELISA | Polyspecific | Low* | High heparin dose confirmation step | In-house assays |
| IgG specific | Intermediate† | GTI Diagnostics, Waukesha, WI (GTI-PF4; HAT; PF4-Enhanced; GTI-IgG) | ||
| High‡ | Hyphen-BioMed, Neuville-Sur-Oise, France (Zymutest HIA IgGAM; Zymutest HIA IgG) | |||
| Diagnostica Stago, Asnières-sur-Seine, France (Asserachrom HIPA) | ||||
| Gen-Probe (Gen-Probe PF4)§ | ||||
| Technoclone GmbH, Vienna, Austria (Technozym) | ||||
| PaGIA | Polyspecific | Low|| | Diamed, Cressier sur Morat, Switzerland (ID-H/PF4 PaGIA) | |
| Intermediate¶ | ||||
| PIFA | Polyspecific | Positive/negative | Akers Biosciences Inc, Thorofare, NJ (HealthTEST) | |
| Lateral flow immunoassay | IgG specific | Positive/negative | Diagnostica Stago (STic EXPERT HIT); Milenia Biotec, Giessen, Germany (Milenia QuickLine HIT) | |
| CLIA | Polyspecific | Low# | Instrumentation Laboratory, Bedford, MA (HemosIL AcuStar HIT-Ab; HemosIL AcuStar HIT-IgG) | |
| IgG specific | Intermediate** | |||
| High†† | ||||
| Latex agglutination assay | Polyspecific | Low‡‡ | Instrumentation Laboratory (HemosIL HIT-Ab) |
| Classes of assays . | Antibody specificity . | Threshold . | Test variation . | Manufacturer (names of tests) . |
|---|---|---|---|---|
| ELISA | Polyspecific | Low* | High heparin dose confirmation step | In-house assays |
| IgG specific | Intermediate† | GTI Diagnostics, Waukesha, WI (GTI-PF4; HAT; PF4-Enhanced; GTI-IgG) | ||
| High‡ | Hyphen-BioMed, Neuville-Sur-Oise, France (Zymutest HIA IgGAM; Zymutest HIA IgG) | |||
| Diagnostica Stago, Asnières-sur-Seine, France (Asserachrom HIPA) | ||||
| Gen-Probe (Gen-Probe PF4)§ | ||||
| Technoclone GmbH, Vienna, Austria (Technozym) | ||||
| PaGIA | Polyspecific | Low|| | Diamed, Cressier sur Morat, Switzerland (ID-H/PF4 PaGIA) | |
| Intermediate¶ | ||||
| PIFA | Polyspecific | Positive/negative | Akers Biosciences Inc, Thorofare, NJ (HealthTEST) | |
| Lateral flow immunoassay | IgG specific | Positive/negative | Diagnostica Stago (STic EXPERT HIT); Milenia Biotec, Giessen, Germany (Milenia QuickLine HIT) | |
| CLIA | Polyspecific | Low# | Instrumentation Laboratory, Bedford, MA (HemosIL AcuStar HIT-Ab; HemosIL AcuStar HIT-IgG) | |
| IgG specific | Intermediate** | |||
| High†† | ||||
| Latex agglutination assay | Polyspecific | Low‡‡ | Instrumentation Laboratory (HemosIL HIT-Ab) |
GTI, Genetic Testing Institute.
≤OD 0.7.
Between OD 0.8 and 1.4.
>OD 1.4.
Technically identical with GTI assay.
Positive/negative.
Titer 2 to 3.
1.0 U/mL.
1.0-2.8 U/mL.
>2.8 U/mL.
>3.85 U/mL.
Methodological quality
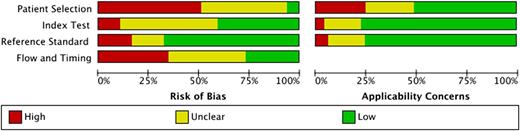
A summary of the methodological quality is shown in Figure 2; the quality of the individual studies is reported in supplemental Figure 1. A low risk of bias in all 4 domains was observed in 2 of 128 evaluation studies only (1.6%).68 The median number of low-risk ratings was 1 (25%). A high risk of bias was assessed in 125 cases (24.4%). Most high-risk ratings were assigned in the domain patient selection (57) and the fewest in index test (11). Most low-risk ratings were found in the reference standard domain (90). In contrast, low concerns regarding applicability were assessed in 276 cases (71.9%). Methodological criteria that were frequently inadequately addressed were (1) prospectively enrolled patients, (2) specified cohort criteria, (3) numbers and reasons for excluded patients, (4) interpretation of reference standard without knowledge of the index test, and (5) clearly described sequence of testing.
Summary of methodological quality. Methodological quality of studies investigating diagnostic accuracy of immunoassays for diagnosis of HIT was assessed using QUADAS-2.29 Detailed results for individual studies are shown in supplemental Figure 1.
Summary of methodological quality. Methodological quality of studies investigating diagnostic accuracy of immunoassays for diagnosis of HIT was assessed using QUADAS-2.29 Detailed results for individual studies are shown in supplemental Figure 1.
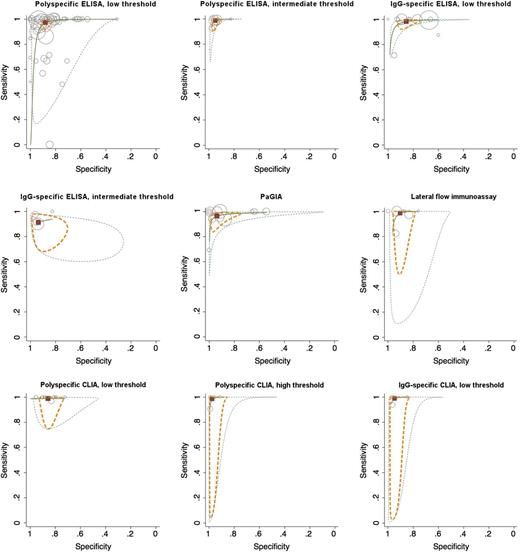
Diagnostic accuracy of immunoassay classes: HSROC and bivariate model
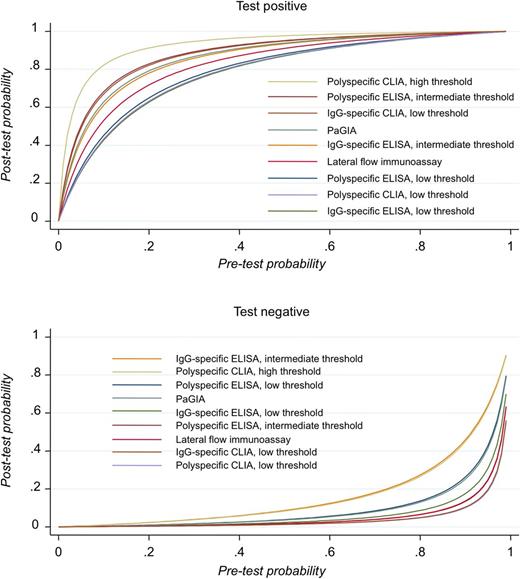
We were able to calculate diagnostic accuracy measures according to the unified model for 9 classes of assays: polyspecific ELISA with low and intermediate threshold, IgG-specific ELISA with low and intermediate threshold, PaGIA, lateral flow immunoassay, polyspecific CLIA with low and high threshold, and IgG-specific CLIA with low threshold. Detailed results are reported in Table 2; Figure 3 illustrates diagnostic accuracy as a hierarchical summary receiver operating characteristics (HSROC) curve. Among the categories that could be analyzed with the unified model, sensitivity was >95% for polyspecific ELISA with low and intermediate threshold, IgG-specific ELISA with low threshold, PaGIA, lateral flow immunoassay, polyspecific CLIA with low and high threshold, and IgG-specific CLIA with low threshold. Specificity was >90% in polyspecific ELISA with intermediate threshold, IgG-specific ELISA with intermediate threshold, PaGIA, lateral flow immunoassay, polyspecific CLIA with high threshold, and IgG-specific CLIA with low threshold. Both criteria were fulfilled in 5 assay classes (polyspecific ELISA with intermediate threshold, PaGIA, lateral flow immunoassay, polyspecific CLIA with high threshold, IgG-specific CLIA with low threshold). A forest plot was generated to compare diagnostic accuracy measures (Figure 4). To illustrate the diagnostic value of the tests at different pretest probabilities (prevalence or probability according to clinical scoring tools), we show posttest probabilities of positive and negative test results in Table 2 and Figure 5 (assuming constant likelihood ratios).
Pooled diagnostic accuracy measures of different classes of immunoassays for diagnosis of HIT
| Type of test . | No. studies; no. patients* . | Statistical model . | Sens. (%) . | Spec. (%) . | LR . | Posttest probability of pos. test result . | Posttest probability of neg. test result . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pos. (95% CI) . | Neg. (95% CI) . | 1% Prev. . | 7% Prev. . | 15% Prev. . | 1% Prev. . | 7% Prev. . | 15% Prev. . | |||||
| Polyspecific ELISA | ||||||||||||
| LT16,19,20,28,39,41,43,44,46,47,49-54,59-61,63,64,68,69,71,74,75,77-79 | 31; 8933 | Unified† | 96.7 (89.7, 99.0) | 86.8 (82.0, 90.5) | 7.3 (5.4-10.0) | 0.04 (0.01- 0.12) | 6.9 (5.1, 9.2) | 35.5 (28.9, 42.9) | 56.3 (48.9, 63.8) | 0.0 (0.0, 0.1) | 0.3 (0.1, 0.9) | 0.7 (0.2, 2.1) |
| IT19,47,61,65,77 | 5; 2334 | Unified† | 98.4 (90.8, 99.7) | 94.9 (90.5, 97.3) | 19.3 (10.4-36.0) | 0.02 (0.00-0.1) | 16.3 (9.5, 26.7) | 59.2 (43.9, 73.0) | 77.3 (64.7, 86.4) | 0.0 (0.0, 0.1) | 0.1 (0.0, 0.7) | 0.4 (0.0, 1.7) |
| HT19,39,47 | 3; 1014 | Fixed effects‡ | 15.0 (14.5, 15.5) | 100 (99.3, 100) | 73.4 (28.2-190.9) | 0.3 (0.2-0.5) | 42.6 (22.2, 65.9) | 84.7 (68.0, 93.5) | 92.8 (83.3, 97.1) | 0.3 (0.2, 0.5) | 2.2 (1.5, 3.6) | 5.0 (3.4, 8.1) |
| IgG-specific ELISA | ||||||||||||
| LT19,20,44,55,63,64,66,69-72,77 | 12; 3116 | Unified† | 98.3 (95.1, 99.4) | 85.4 (78.2, 90.6) | 6.7 (4.5-10.2) | 0.02 (0.01-0.05) | 6.3 (4.3, 9.3) | 33.5 (25.3, 43.4) | 54.2 (44.3, 64.3) | 0.0 (0.0, 0.1) | 0.2 (0.1, 0.4) | 0.4 (0.2, 0.9) |
| IT19,20,77 | 4; 2545 | Unified† | 91.2 (86.2, 94.5) | 93.5 (89.1-96.2) | 14.1 (8.1-24.5) | 0.09 (0.05-0.15) | 12.5 (7.6, 19.8) | 51.5 (38.0, 64.8) | 71.3 (58.9, 81.2) | 0.1 (0.1, 0.2) | 0.7 (0.4, 1.1) | 1.6 (1.0, 2.6) |
| HT19 | 2; 1958 | Fixed effects‡ | 60.9 (59.7, 62.1) | 99.4 (97.6-100) | 97.0 (53.0-177.6) | 0.4 (0.3-0.5) | 49.5 (34.9, 64.2) | 88.0 (80.0, 93.0) | 94.5 (90.3, 96.9) | 0.4 (0.3, 0.5) | 2.9 (2.2, 3.6) | 6.6 (5.0, 8.1) |
| PaGIA | ||||||||||||
| LT20,36,42,45,46,53,58,64,68,70,72,75,79 | 12; 2205 | Unified† | 96.5 (89.8, 98.9) | 93.7 (83.1, 97.8) | 15.3 (5.5, 42.3) | 0.04 (0.01, 0.11) | 13.4 (5.3, 29.9) | 53.5 (29.3, 76.1) | 73.0 (49.3, 88.2) | 0.0 (0.0, 0.1) | 0.3 (0.1, 0.8) | 0.7 (0.2, 1.8) |
| IT65 | 1; 1291 | None§ | 98.9 | 95.9 | 24.1 | 0.01 | 19.6 | 64.5 | 81.0 | 0.0 | 0.1 | 0.2 |
| Lateral flow immunoassay54-56,58,66,70,72 | 7; 1163 | Unified† | 98.4 (85.3, 99.9) | 90.3 (84.4, 94.1) | 10.1 (6.2, 16.5) | 0.02 (0.00, 0.18) | 9.3 (5.9, 14.3) | 43.3 (31.9, 55.4) | 64.2 (52.4, 74.5) | 0.0 (0.0, 0.2) | 0.1 (0.0, 1.3) | 0.3 (0.0, 3.0) |
| PIFA28 | 1; 88 | None§ | 0.0 | 70.1 | 2.3 | 0.5 | 2.3 | 14.8 | 28.9 | 0.5 | 3.6 | 8.1 |
| Latex agglutination assay37 | 1; 119 | None§ | 100.0 | 84.3 | 3.7 | 0.0 | 3.6 | 21.8 | 39.5 | 0.0 | 0.0 | 0.0 |
| Polyspecific CLIA | ||||||||||||
| LT37,38,57,61,62,77 | 7; 1008 | Unified† | 98.9 (92.7, 99.8) | 85.6 (79.3, 90.3) | 6.9 (4.7-10.0) | 0.01 (0.00-0.09) | 6.5 (4.5, 9.2) | 34.2 (26.1, 42.9) | 54.9 (45.4, 63.8) | 0.0 (0.0, 0.1) | 0.1 (0.0, 0.7) | 0.2 (0.0, 1.6) |
| IT37 | 2; 448 | Fixed effects‡ | 97.9 (94.6, 100.0) | 93.1 (90.4, 95.8) | 13.5 (9.5-18.9) | 0.0 (0.0-0.1) | 12.0 (8.8, 16.0) | 50.4 (41.7, 58.7) | 70.4 (62.6, 76.9) | 0.0 (0.0, 0.1) | 0.0 (0.0, 0.7) | 0.0 (0.0, 1.7) |
| HT37,61,62,77 | 5; 755 | Unified† | 98.3 (69.5, 99.9) | 97.5 (94.4, 98.9) | 39.5 (17.5-89.2) | 0.0 (0.0-0.40) | 28.5 (15.0, 47.6) | 74.8 (56.8, 87.1) | 87.5 (75.5, 94.1) | 0.0 (0.0, 0.4) | 0.1 (0.0, 2.9) | 0.2 (0.0, 6.6) |
| IgG-specific CLIA | ||||||||||||
| LT37,57,61,77 | 5; 741 | Unified† | 98.8 (69.2, 100.0) | 94.6 (90.7, 96.9) | 18.3 (10.6-31.5) | 0.01 (0.00- 0.40) | 15.6 (9.7, 24.1) | 57.9 (44.4, 70.3) | 76.3 (65.2, 84.8) | 0.0 (0.0, 0.4) | 0.1 (0.0, 3.2) | 0.2 (0.0, 7.2) |
| IT37 | 2; 448 | Fixed effects‡ | 78.6 (75.9, 81.2) | 98.7 (94.6, 100) | 42.3 (20.1-88.7) | 0.2 (0.1-0.3) | 29.9 (16.9, 47.3) | 76.1 (60.2, 87.0) | 88.2 (78.0, 94.0) | 0.2 (0.1, 0.3) | 1.5 (0.7, 2.2) | 3.4 (1.7, 5.0) |
| HT37,61 | 3; 552 | Fixed effects‡ | 74.2 (71.9, 76.5) | 99.1 (95.4, 100) | 47.8 (23.2-98.7) | 0.2 (0.1-0.4) | 32.6 (19.0, 49.9) | 78.3 (63.6, 88.1) | 89.4 (80.4, 94.6) | 0.2 (0.1, 0.4) | 1.5 (0.7, 2.9) | 3.4 (1.7, 6.6) |
| High-dose heparin confirmation step | ||||||||||||
| Polyspecific ELISA, LT21,40 | 2; 116 | Fixed effects‡ | 99.4 (94.7, 100)|| | 85.8 (83.1, 88.5) | 2.7 (1.6-4.7) | 0.3 (0.1-1.3) | 2.7 (1.6, 4.5) | 16.9 (10.7, 26.1) | 32.3 (22.0, 45.3) | 0.3 (0.1, 1.3) | 2.2 (0.7, 8.9) | 5.0 (1.7, 18.7) |
| IgG-specific ELISA, LT21 | 3; 1459 | Fixed effects‡ | 88.3 (87.0, 89.8) | 90.7 (89.2, 92.1) | 9.3 (7.8-11.1) | 0.1 (0.1-0.2) | 8.6 (7.3, 10.1) | 41.2 (37.0, 45.5) | 62.2 (57.9, 66.2) | 0.1 (0.1, 0.2) | 0.7 (0.7, 1.5) | 1.7 (1.7, 3.4) |
| IgG-specific ELISA, IT21 | 2; 1000 | Fixed effects‡ | 69.7 (68.2, 71.3) | 96.2 (94.7, 97.8) | 18.2 (12.8-25.8) | 0.3 (0.2-0.4) | 15.5 (11.4, 20.7) | 57.8 (49.1, 66.0) | 76.3 (69.3, 82.0) | 0.3 (0.2, 0.4) | 2.2 (1.5, 2.9) | 5.0 (3.4, 6.6) |
| Type of test . | No. studies; no. patients* . | Statistical model . | Sens. (%) . | Spec. (%) . | LR . | Posttest probability of pos. test result . | Posttest probability of neg. test result . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pos. (95% CI) . | Neg. (95% CI) . | 1% Prev. . | 7% Prev. . | 15% Prev. . | 1% Prev. . | 7% Prev. . | 15% Prev. . | |||||
| Polyspecific ELISA | ||||||||||||
| LT16,19,20,28,39,41,43,44,46,47,49-54,59-61,63,64,68,69,71,74,75,77-79 | 31; 8933 | Unified† | 96.7 (89.7, 99.0) | 86.8 (82.0, 90.5) | 7.3 (5.4-10.0) | 0.04 (0.01- 0.12) | 6.9 (5.1, 9.2) | 35.5 (28.9, 42.9) | 56.3 (48.9, 63.8) | 0.0 (0.0, 0.1) | 0.3 (0.1, 0.9) | 0.7 (0.2, 2.1) |
| IT19,47,61,65,77 | 5; 2334 | Unified† | 98.4 (90.8, 99.7) | 94.9 (90.5, 97.3) | 19.3 (10.4-36.0) | 0.02 (0.00-0.1) | 16.3 (9.5, 26.7) | 59.2 (43.9, 73.0) | 77.3 (64.7, 86.4) | 0.0 (0.0, 0.1) | 0.1 (0.0, 0.7) | 0.4 (0.0, 1.7) |
| HT19,39,47 | 3; 1014 | Fixed effects‡ | 15.0 (14.5, 15.5) | 100 (99.3, 100) | 73.4 (28.2-190.9) | 0.3 (0.2-0.5) | 42.6 (22.2, 65.9) | 84.7 (68.0, 93.5) | 92.8 (83.3, 97.1) | 0.3 (0.2, 0.5) | 2.2 (1.5, 3.6) | 5.0 (3.4, 8.1) |
| IgG-specific ELISA | ||||||||||||
| LT19,20,44,55,63,64,66,69-72,77 | 12; 3116 | Unified† | 98.3 (95.1, 99.4) | 85.4 (78.2, 90.6) | 6.7 (4.5-10.2) | 0.02 (0.01-0.05) | 6.3 (4.3, 9.3) | 33.5 (25.3, 43.4) | 54.2 (44.3, 64.3) | 0.0 (0.0, 0.1) | 0.2 (0.1, 0.4) | 0.4 (0.2, 0.9) |
| IT19,20,77 | 4; 2545 | Unified† | 91.2 (86.2, 94.5) | 93.5 (89.1-96.2) | 14.1 (8.1-24.5) | 0.09 (0.05-0.15) | 12.5 (7.6, 19.8) | 51.5 (38.0, 64.8) | 71.3 (58.9, 81.2) | 0.1 (0.1, 0.2) | 0.7 (0.4, 1.1) | 1.6 (1.0, 2.6) |
| HT19 | 2; 1958 | Fixed effects‡ | 60.9 (59.7, 62.1) | 99.4 (97.6-100) | 97.0 (53.0-177.6) | 0.4 (0.3-0.5) | 49.5 (34.9, 64.2) | 88.0 (80.0, 93.0) | 94.5 (90.3, 96.9) | 0.4 (0.3, 0.5) | 2.9 (2.2, 3.6) | 6.6 (5.0, 8.1) |
| PaGIA | ||||||||||||
| LT20,36,42,45,46,53,58,64,68,70,72,75,79 | 12; 2205 | Unified† | 96.5 (89.8, 98.9) | 93.7 (83.1, 97.8) | 15.3 (5.5, 42.3) | 0.04 (0.01, 0.11) | 13.4 (5.3, 29.9) | 53.5 (29.3, 76.1) | 73.0 (49.3, 88.2) | 0.0 (0.0, 0.1) | 0.3 (0.1, 0.8) | 0.7 (0.2, 1.8) |
| IT65 | 1; 1291 | None§ | 98.9 | 95.9 | 24.1 | 0.01 | 19.6 | 64.5 | 81.0 | 0.0 | 0.1 | 0.2 |
| Lateral flow immunoassay54-56,58,66,70,72 | 7; 1163 | Unified† | 98.4 (85.3, 99.9) | 90.3 (84.4, 94.1) | 10.1 (6.2, 16.5) | 0.02 (0.00, 0.18) | 9.3 (5.9, 14.3) | 43.3 (31.9, 55.4) | 64.2 (52.4, 74.5) | 0.0 (0.0, 0.2) | 0.1 (0.0, 1.3) | 0.3 (0.0, 3.0) |
| PIFA28 | 1; 88 | None§ | 0.0 | 70.1 | 2.3 | 0.5 | 2.3 | 14.8 | 28.9 | 0.5 | 3.6 | 8.1 |
| Latex agglutination assay37 | 1; 119 | None§ | 100.0 | 84.3 | 3.7 | 0.0 | 3.6 | 21.8 | 39.5 | 0.0 | 0.0 | 0.0 |
| Polyspecific CLIA | ||||||||||||
| LT37,38,57,61,62,77 | 7; 1008 | Unified† | 98.9 (92.7, 99.8) | 85.6 (79.3, 90.3) | 6.9 (4.7-10.0) | 0.01 (0.00-0.09) | 6.5 (4.5, 9.2) | 34.2 (26.1, 42.9) | 54.9 (45.4, 63.8) | 0.0 (0.0, 0.1) | 0.1 (0.0, 0.7) | 0.2 (0.0, 1.6) |
| IT37 | 2; 448 | Fixed effects‡ | 97.9 (94.6, 100.0) | 93.1 (90.4, 95.8) | 13.5 (9.5-18.9) | 0.0 (0.0-0.1) | 12.0 (8.8, 16.0) | 50.4 (41.7, 58.7) | 70.4 (62.6, 76.9) | 0.0 (0.0, 0.1) | 0.0 (0.0, 0.7) | 0.0 (0.0, 1.7) |
| HT37,61,62,77 | 5; 755 | Unified† | 98.3 (69.5, 99.9) | 97.5 (94.4, 98.9) | 39.5 (17.5-89.2) | 0.0 (0.0-0.40) | 28.5 (15.0, 47.6) | 74.8 (56.8, 87.1) | 87.5 (75.5, 94.1) | 0.0 (0.0, 0.4) | 0.1 (0.0, 2.9) | 0.2 (0.0, 6.6) |
| IgG-specific CLIA | ||||||||||||
| LT37,57,61,77 | 5; 741 | Unified† | 98.8 (69.2, 100.0) | 94.6 (90.7, 96.9) | 18.3 (10.6-31.5) | 0.01 (0.00- 0.40) | 15.6 (9.7, 24.1) | 57.9 (44.4, 70.3) | 76.3 (65.2, 84.8) | 0.0 (0.0, 0.4) | 0.1 (0.0, 3.2) | 0.2 (0.0, 7.2) |
| IT37 | 2; 448 | Fixed effects‡ | 78.6 (75.9, 81.2) | 98.7 (94.6, 100) | 42.3 (20.1-88.7) | 0.2 (0.1-0.3) | 29.9 (16.9, 47.3) | 76.1 (60.2, 87.0) | 88.2 (78.0, 94.0) | 0.2 (0.1, 0.3) | 1.5 (0.7, 2.2) | 3.4 (1.7, 5.0) |
| HT37,61 | 3; 552 | Fixed effects‡ | 74.2 (71.9, 76.5) | 99.1 (95.4, 100) | 47.8 (23.2-98.7) | 0.2 (0.1-0.4) | 32.6 (19.0, 49.9) | 78.3 (63.6, 88.1) | 89.4 (80.4, 94.6) | 0.2 (0.1, 0.4) | 1.5 (0.7, 2.9) | 3.4 (1.7, 6.6) |
| High-dose heparin confirmation step | ||||||||||||
| Polyspecific ELISA, LT21,40 | 2; 116 | Fixed effects‡ | 99.4 (94.7, 100)|| | 85.8 (83.1, 88.5) | 2.7 (1.6-4.7) | 0.3 (0.1-1.3) | 2.7 (1.6, 4.5) | 16.9 (10.7, 26.1) | 32.3 (22.0, 45.3) | 0.3 (0.1, 1.3) | 2.2 (0.7, 8.9) | 5.0 (1.7, 18.7) |
| IgG-specific ELISA, LT21 | 3; 1459 | Fixed effects‡ | 88.3 (87.0, 89.8) | 90.7 (89.2, 92.1) | 9.3 (7.8-11.1) | 0.1 (0.1-0.2) | 8.6 (7.3, 10.1) | 41.2 (37.0, 45.5) | 62.2 (57.9, 66.2) | 0.1 (0.1, 0.2) | 0.7 (0.7, 1.5) | 1.7 (1.7, 3.4) |
| IgG-specific ELISA, IT21 | 2; 1000 | Fixed effects‡ | 69.7 (68.2, 71.3) | 96.2 (94.7, 97.8) | 18.2 (12.8-25.8) | 0.3 (0.2-0.4) | 15.5 (11.4, 20.7) | 57.8 (49.1, 66.0) | 76.3 (69.3, 82.0) | 0.3 (0.2, 0.4) | 2.2 (1.5, 2.9) | 5.0 (3.4, 6.6) |
HT, high threshold; IT, intermediate threshold; LR, likelihood ratio; LT, low threshold; neg., negative; pos., positive; prev., prevalence; sens., sensitivity; spec., specificity.
Studies conducted in the same study population were included only once to maintain independency of observations.
In tests with <4 studies available, summary estimates were calculated using a fixed-effects model (Mantel-Haenszel method); however, the results of this analysis must be interpreted with caution because it is not accepted as a valid statistical technique in this situation.
Only 1 study available.
Result of fixed-effects model appears invalid because the favorable sensitivity depends on 1 study in which the only 1 HIT case was detected.67
Sensitivity and specificity of immunoassay classes for diagnosis of HIT. Results of an HSROC model are reported showing the summary point (red quadrant), a 95% CI region (dashed yellow line), and 95% prediction region (dashed green line), illustrating a confidence region for the true sensitivity and specificity in a possible future study. Results of primary studies are shown as circles; the sample size of the studies is represented by the size of the circle.
Sensitivity and specificity of immunoassay classes for diagnosis of HIT. Results of an HSROC model are reported showing the summary point (red quadrant), a 95% CI region (dashed yellow line), and 95% prediction region (dashed green line), illustrating a confidence region for the true sensitivity and specificity in a possible future study. Results of primary studies are shown as circles; the sample size of the studies is represented by the size of the circle.
Diagnostic accuracy of different immunoassay classes as characterized by positive and negative likelihood ratios. Likelihood ratios (LR) are powerful measures describing how many times more (or less) likely a test result is in patients with the disease in contrast to patients without the disease.80 In the context of HIT, a test with a +LR above 10 (corresponding to a specificity of 90%) and a –LR below 0.05 (corresponding to a sensitivity of 95%) is considered favorable.
Diagnostic accuracy of different immunoassay classes as characterized by positive and negative likelihood ratios. Likelihood ratios (LR) are powerful measures describing how many times more (or less) likely a test result is in patients with the disease in contrast to patients without the disease.80 In the context of HIT, a test with a +LR above 10 (corresponding to a specificity of 90%) and a –LR below 0.05 (corresponding to a sensitivity of 95%) is considered favorable.
Probability of HIT after conducting an immunoassay at varying pretest probabilities. Physicians are interested to know how likely a disease is with a positive test result and how unlikely it is with a negative test results. The posttest probability depends largely on the pretest probability (corresponding to the prevalence or the result of a clinical scoring system), but also on the likelihood ratios of the test.
Probability of HIT after conducting an immunoassay at varying pretest probabilities. Physicians are interested to know how likely a disease is with a positive test result and how unlikely it is with a negative test results. The posttest probability depends largely on the pretest probability (corresponding to the prevalence or the result of a clinical scoring system), but also on the likelihood ratios of the test.
Approximate diagnostic accuracy of immunoassay classes: fixed-effects model
Because of a limited number of studies, we were unable to calculate diagnostic accuracy measures using the unified model in 9 tests: polyspecific ELISA and IgG-specific ELISA with high threshold, polyspecific ELISA and IgG-specific ELISA with high-dose heparin confirmation step (low and intermediate threshold), PaGIA with intermediate threshold, polyspecific CLIA with intermediate threshold, and IgG-specific CLIA with intermediate and high threshold. To give an impression of the diagnostic accuracy in these tests, we calculated diagnostic accuracy measures using a fixed-effects model (if possible) and report the results in Table 2. Sensitivity was below 95% in all of the previously mentioned tests except polyspecific ELISA with low threshold and high heparin dose confirmation step.
Diagnostic accuracy of ELISA assays by manufacturer
We were able to calculate diagnostic accuracy measures for the individual ELISA manufacturers by means of the unified model in 8 assays and by the use of a fixed-effects model in 2 assays. No pooling was possible in 4 assays with a unique evaluation study. Results are reported in Table 3. In most of the assays, diagnostic accuracy corresponds to the general characteristic of the class (sensitivity above or below 95%; specificity above or below 90%). However, some assays appear to be inferior (eg, IgG-specific Asserachrom assay with a low threshold). Diagnostic accuracy was superior to corresponding class in case of IgG-specific Genetic Testing Institute-ELISA with low threshold (sensitivity, 99.6%, 95% confidence interval [CI] 22.7-100.0; specificity, 89.9%, 95% CI 86.2-92.6).
Diagnostic accuracy of different ELISA assays by manufacturer
| Assay . | No. studies . | No. patients . | Statistical model . | Sensitivity (95% CI) . | Specificity (95% CI) . | Positive LR (95% CI) . | Negative LR (95% CI) . |
|---|---|---|---|---|---|---|---|
| Polyspecific ELISA, low threshold | |||||||
| Pooled estimates | 31* | 8933* | Unified | 96.7 (89.7-99.0) | 86.8 (82.0-90.5) | 7.3 (5.4-10.0) | 0.04 (0.01-0.12) |
| GTI19,20,28,41,46,47,50,54,61,68,74,75,79 † | 13 | 3804 | Unified | 99.9 (90.9-100.0) | 87.4 (79.2-92.7) | 7.9 (4.7-13.4) | 0.001 (<0.001-0.11) |
| Zymutest63,69,77 | 3 | 295 | Fixed effects‡ | 99.5 (94.8-100.0) | 77.6 (72.9-82.3) | 4.1 (3.1-5.5) | 0.05 (0.01-0.12) |
| Asserachrom39,45,46,49-51,53,59,60,63,64,71,75,78,79 | 15 | 2621 | Unified | 92.7 (73.6-98.3) | 87.3 (79.9-92.3) | 7.3 (4.6-11.7) | 0.08 (0.02-0.34) |
| In-house assay16,43,48,52,79 § | 5 | 2673 | Unified | 93.2 (76.3-98.3) | 92.6 (90.1-94.0) | 12.6 (9.7-16.3) | 0.07 (0.02-0.28) |
| Polyspecific ELISA, intermediate threshold | |||||||
| Pooled estimates* | 5* | 2334* | Unified | 98.6 (95.2-99.6) | 94.8 (92.2-96.5) | 18.8 (12.7-27.9) | 0.01 (0.004-0.05) |
| GTI19,47,61,65 | 5 | 2646 | Unified | 97.4 (92.4-99.1) | 95.9 (94.3-97.0) | 23.6 (17.2-32.4) | 0.03 (0.009-0.08) |
| Zymutest77 | 1 | 87 | — | 100.0 | 82.5 | 5.7 | 0.00 |
| Asserachrom65 | 1 | 1291 | — | 99.6 | 93.6 | 15.6 | 0.01 |
| IgG-specific ELISA, low threshold | |||||||
| Pooled estimates* | 12* | 3116* | Unified | 98.3 (95.1-99.4) | 85.4 (78.2-90.6) | 6.7 (4.5-10.2) | 0.02 (0.01-0.05) |
| GTI55,63,64,70 | 4 | 639 | Unified | 99.6 (22.7-100.0) | 89.9 (86.2-92.6) | 9.8 (7.2-13.4) | 0.004 (<0.001-3.7) |
| Zymutest44,63,69,70,77 | 5 | 829 | Unified | 99.2 (86.4-100.0) | 85.8 (77.1-91.5) | 7.0 (4.3-11.4) | 0.008 (<0.001-0.18) |
| Asserachrom66,71 | 2 | 160 | Fixed effects‡ | 72.0 (68.4-75.5) | 93.8 (90.3-97.4) | 3.6 (2.04-5.6) | 0.27 (0.10-0.72) |
| Technozym72 | 1 | 41 | — | 100.0 | 0.92 | 12.7 | 0.00 |
| In-house assay19,20 § | 4 | 2958 | Unified | 99.2 (96.4-99.8) | 83.5 (71.5-91.0) | 6.0 (3.4-10.7) | 0.009 (0.002-0.04) |
| IgG-specific ELISA, intermediate threshold | |||||||
| Pooled estimates* | 4* | 2545* | Unified | 91.2 (86.3-94.5) | 93.5 (89.1-96.2) | 14.1 (8.1-24.5) | 0.09 (0.06-0.15) |
| Zymutest77 | 1 | 87 | — | 100.0 | 82.5 | 5.7 | 0.00 |
| In-house assay19,20 § | 5 | 4416 | Unified | 89.8 (82.7-94.2) | 96.0 (94.5-97.1) | 22.5 (16.3-31.2) | 0.10 (0.06-0.18) |
| Assay . | No. studies . | No. patients . | Statistical model . | Sensitivity (95% CI) . | Specificity (95% CI) . | Positive LR (95% CI) . | Negative LR (95% CI) . |
|---|---|---|---|---|---|---|---|
| Polyspecific ELISA, low threshold | |||||||
| Pooled estimates | 31* | 8933* | Unified | 96.7 (89.7-99.0) | 86.8 (82.0-90.5) | 7.3 (5.4-10.0) | 0.04 (0.01-0.12) |
| GTI19,20,28,41,46,47,50,54,61,68,74,75,79 † | 13 | 3804 | Unified | 99.9 (90.9-100.0) | 87.4 (79.2-92.7) | 7.9 (4.7-13.4) | 0.001 (<0.001-0.11) |
| Zymutest63,69,77 | 3 | 295 | Fixed effects‡ | 99.5 (94.8-100.0) | 77.6 (72.9-82.3) | 4.1 (3.1-5.5) | 0.05 (0.01-0.12) |
| Asserachrom39,45,46,49-51,53,59,60,63,64,71,75,78,79 | 15 | 2621 | Unified | 92.7 (73.6-98.3) | 87.3 (79.9-92.3) | 7.3 (4.6-11.7) | 0.08 (0.02-0.34) |
| In-house assay16,43,48,52,79 § | 5 | 2673 | Unified | 93.2 (76.3-98.3) | 92.6 (90.1-94.0) | 12.6 (9.7-16.3) | 0.07 (0.02-0.28) |
| Polyspecific ELISA, intermediate threshold | |||||||
| Pooled estimates* | 5* | 2334* | Unified | 98.6 (95.2-99.6) | 94.8 (92.2-96.5) | 18.8 (12.7-27.9) | 0.01 (0.004-0.05) |
| GTI19,47,61,65 | 5 | 2646 | Unified | 97.4 (92.4-99.1) | 95.9 (94.3-97.0) | 23.6 (17.2-32.4) | 0.03 (0.009-0.08) |
| Zymutest77 | 1 | 87 | — | 100.0 | 82.5 | 5.7 | 0.00 |
| Asserachrom65 | 1 | 1291 | — | 99.6 | 93.6 | 15.6 | 0.01 |
| IgG-specific ELISA, low threshold | |||||||
| Pooled estimates* | 12* | 3116* | Unified | 98.3 (95.1-99.4) | 85.4 (78.2-90.6) | 6.7 (4.5-10.2) | 0.02 (0.01-0.05) |
| GTI55,63,64,70 | 4 | 639 | Unified | 99.6 (22.7-100.0) | 89.9 (86.2-92.6) | 9.8 (7.2-13.4) | 0.004 (<0.001-3.7) |
| Zymutest44,63,69,70,77 | 5 | 829 | Unified | 99.2 (86.4-100.0) | 85.8 (77.1-91.5) | 7.0 (4.3-11.4) | 0.008 (<0.001-0.18) |
| Asserachrom66,71 | 2 | 160 | Fixed effects‡ | 72.0 (68.4-75.5) | 93.8 (90.3-97.4) | 3.6 (2.04-5.6) | 0.27 (0.10-0.72) |
| Technozym72 | 1 | 41 | — | 100.0 | 0.92 | 12.7 | 0.00 |
| In-house assay19,20 § | 4 | 2958 | Unified | 99.2 (96.4-99.8) | 83.5 (71.5-91.0) | 6.0 (3.4-10.7) | 0.009 (0.002-0.04) |
| IgG-specific ELISA, intermediate threshold | |||||||
| Pooled estimates* | 4* | 2545* | Unified | 91.2 (86.3-94.5) | 93.5 (89.1-96.2) | 14.1 (8.1-24.5) | 0.09 (0.06-0.15) |
| Zymutest77 | 1 | 87 | — | 100.0 | 82.5 | 5.7 | 0.00 |
| In-house assay19,20 § | 5 | 4416 | Unified | 89.8 (82.7-94.2) | 96.0 (94.5-97.1) | 22.5 (16.3-31.2) | 0.10 (0.06-0.18) |
Studies conducted in the same study population were included only once to maintain independency of observations.
Including technically identical Gen-Probe assay.
In tests with <4 studies available, summary estimates were calculated using a fixed-effects model (Mantel-Haenszel method; to be interpreted with caution).
Application of pooled “in-house assay” data to individual tests must be done with caution because these tests are constructed differently (every assay must be evaluated individually).
Potential sources of heterogeneity and bias
For the first sensitivity analysis, calculations were repeated in studies with cohort designs only (lower risk of bias). Only minor differences were observed (range, 0.0%-0.5% for sensitivity; 0.0%-3.6% for specificity), the results are shown in supplemental Table 2. In the second sensitivity analysis, data of studies that used SRA/HIPA as a reference standard were pooled only (supplemental Table 5) and compared with results from all studies. Differences ranged between 0.2% and 1.7% for sensitivity and 0.7% and 1.5% for specificity. Third, we compared data of different tests obtained within the same study population using the same reference standard. Supplemental Table 4 shows the results of the 2 populations with the most tests evaluated. The results of the individual studies and the differences between different categories (thresholds, antibody specificity, class of assays) correspond very well with the pooled data. Supplemental Table 3 shows results of a meta-regression analysis exploring the impact of the reference standard used, setting, and specimen material on diagnostic accuracy. No significant results were observed for any of the previously mentioned variables.
In general, neither sensitivity analyses nor meta-regression recognized any characteristic of study design as possible source of heterogeneity and bias.
Discussion
Key findings
Important differences exist between classes of immunoassays, type of antibodies, thresholds, and application of confirmation step; only 5 tests appear optimal for clinical use. Relevant variations were found even between manufacturers of ELISA assays. In general, methodological quality of primary studies was moderate. Nevertheless, the results were consistent and no differences were found in sensitivity analyses.
Strengths and limitations
The present investigation is the first systematic review and meta-analysis focusing on immunoassays for diagnosis of HIT. We conducted a comprehensive literature search to retrieve the published evidence, applied strict inclusion criteria, assessed the methodological quality of the studies systematically, and used an appropriate meta-analytic technique to pool the existing data. Thus, we were able to compare diagnostic accuracy measures between classes of assays, manufacturers, antibody specificities, thresholds, and the application of high-dose heparin confirmation step.
Our study has several limitations. First, methodological quality of primary studies was only moderate in general. Even though we cannot fully exclude that this might have influenced our results, we estimate the risk to be low: (1) applying strict inclusion criteria, we excluded 94 publications because of low quality; (2) sensitivity analysis did not show relevant differences if compared with high-quality publications only; and (3) the results were consistent across subgroups. Second, the number of available studies was limited in certain categories of tests, restricting the analysis to a fixed-effects meta-analytic model only (Tables 2 and 3). The results of this analysis must be interpreted with caution because it is not accepted as a valid statistical technique in this situation. The numbers of studies and cases are even more limited in certain categories. For example, the favorable sensitivity of polyspecific ELISA with low threshold and high heparin dose confirmation step largely depends on 1 study in which the only 1 HIT case was detected (corresponding to a sensitivity of 100%).67 The number of studies and patients is limited even for the tests most investigated, leading to a restricted precision with wide CIs. For example, expected differences between polyspecific and IgG-specific ELISA in terms of sensitivity and specificity could not be reproduced in every subgroup (Table 2). And third, several different reference standard tests have been used in evaluation studies and we cannot fully preclude that this might have influenced the results of our investigation. However, we expect the risk to be low for several reasons: (1) a sensitivity analysis comparing diagnostic accuracy measures obtained with 1 of the accepted gold standard SRA/HIPA only, and all results did not find any difference; (2) meta-regression analysis revealed no difference in diagnostic odds ratio if SRA/HIPA were compared with other defined reference standard tests; (3) we applied strict inclusion criteria requiring a clear definition of reference standard tests; and (4) in the majority of studies (95/128), either SRA/HIPA or a combination of SRA/HIPA with clinical criteria was used.
One may argue that a systematic review of diagnostic tests for HIT should focus on sensitivities and specificities. Indeed, sensitivity and specificity are well-known (even though often misinterpreted) measures of diagnostic accuracy, and reporting them is essential. However, the important question in clinical practice is how a particular test result predicts disease, and sensitivities/specificities cannot answer this question. Predictive values are often used, but these measures depend on the prevalence of the disease, and generalization beyond a study population is difficult.80 Likelihood ratios are powerful diagnostic accuracy measures because they are assumed to be independent from prevalence, they express how many times more (or less) likely a test result is in patients with the disease in contrast to patients without the disease, and they can be used to calculate posttest probabilities. In addition, differences in likelihood ratios between tests can be seen clearly in a forest plot. In contrast, it is hard to read a forest plot with estimates of sensitivities or specificities along with CIs when most data are between 90% and 100% (ceiling effect). Following these considerations, we decided to report all sensitivities/specificities in a table and likelihood ratios as a forest plot.
Implications for clinical practice
With the present investigation, we provide aggregated diagnostic accuracy measures for all frequently applied immunoassays used to diagnose HIT. Our results will raise clinicians’ awareness for the characteristics of tests they are confronted with. Using the posttest probabilities provided in Table 2 and Figure 5, clinicians may be able to estimate the probability of HIT in individual patients. If used in combination with clinical scoring systems (which assess the pretest probability), they might substantially improve care in patients with suspected HIT. In addition, laboratory managers can use our results to compare test characteristics directly and base decisions about which test to implement on a sound scientific background.
Implications for future research
No antibody test is both 100% sensitive and specific, and such a test is not expected in the near future. Functional assays are rarely available in a timely fashion. Clinical scoring systems such as the 4Ts score could help identifying patients with a low pretest probability and might reduce the number of “overtreated” patients.12 However, there are several unsolved issues associated with the 4Ts score.12-14 Thus, we will always miss a fraction of patients with HIT, or treat a number of non-HIT patients. Both situations are dangerous. A promising concept to overcome these limitations might be a subsequent or parallel determination of different tests and estimation of posttest probabilities using nomograms. We suggest researchers focus on diagnostic algorithms to improve diagnostic procedures in this vulnerable group of patients.
Conclusions
Important differences exist between immunoassays for diagnosis of HIT, in particular with regard to different classes of assays, antibody specificities, manufacturers, thresholds, and the application of the high-dose heparin confirmation step. Only 5 tests have a high sensitivity combined with a high specificity: polyspecific ELISA with an intermediate threshold (Genetic Testing Institute, Asserachrom), PaGIA, lateral flow immunoassay, polyspecific CLIA with a high threshold, and IgG-specific CLIA with a low threshold. Diagnostic accuracy appears to be inadequate in tests with high thresholds (ELISA, IgG-specific CLIA), combination of IgG specificity and intermediate thresholds (ELISA, CLIA), high-dose heparin confirmation step (ELISA), PIFA, and IgG-specific Asserachrom assay at low threshold. Clinicians should be aware of the pretest probability as well as the diagnostic characteristics of the tests they are confronted with. Investigations focused on diagnostic algorithms combining different tests might further improve care in patients with suspected HIT.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Prof Greinacher, Greifswald (Germany) for his valuable comments on an early version of this manuscript.
This study was supported by Research Fund Haematology Luzerner Kantonsspital.
Authorship
Contribution: M.N. developed the protocol, conducted the literature search, assessed methodological quality, performed data extraction, conducted the statistical analysis, and wrote the manuscript; L.M.B. developed the analysis plan and reviewed the analysis; H.t.C. reviewed the protocol and the manuscript; A.t.C.-H. reviewed the protocol, assessed methodological quality, performed data extraction, and reviewed the manuscript. All authors approved the final version of the manuscript.
Conflict-of-interest disclosure: M.N. has received research grants or lecture fees from Pfizer, Bayer, CSL Behring, Sanofi-Aventis, Roche Diagnostics, and Axonlab. H.t.C. is a Fellow of the Gutenberg Forschungscolleg, Gutenberg University, Mainz, Germany. The remaining authors declare no competing financial interests.
Correspondence: Michael Nagler, Department of Haematology and Central Haematology Laboratory, Inselspital University Hospital, 3010 Bern, Switzerland; e-mail: michael.nagler@insel.ch