Key Points
IKKβ, independently of NF-κB, regulates the stability and transcriptional activity of GLI1 oncogene.
Combined inhibition of IKKβ and GLI1 activities synergistically decreases DLBCL cell viability in vivo and in vitro.
Abstract
GLI1 oncogene has been implicated in the pathobiology of several neoplasms including diffuse large B-cell lymphoma (DLBCL). However, mechanisms underlying GLI1-increased activity in DLBCL are poorly characterized. Herein, we demonstrate that IKKβ phosphorylates GLI1 in DLBCL. IKKβ activation increased GLI1 protein levels and transcriptional activity, whereas IKKβ silencing decreased GLI1 levels and transcriptional activity. Tumor necrosis factor-α (TNFα) mediated IKKβ activation–impaired GLI1 binding with the E3 ubiquitin ligase-ITCH, leading to decreased K48-linked ubiquitination/degradation of GLI1. We found 8 IKKβ-dependent phosphorylation sites that mediate GLI1 stability. Mutating or deleting these residues facilitated GLI1-ITCH interaction and decreased the protective effect of TNFα on GLI1 stability. IKKβ-GLI1 crosstalk is significant because combined inhibition of both molecules resulted in synergistic suppression of DLBCL viability in vivo and in vitro. By linking IKKβ-mediated nuclear factor-κB activity with GLI1, we identified a crosstalk between these 2 pathways that can inform the design of novel therapeutic strategies in DLBCL.
Introduction
GLI1 is a transcription factor that regulates gene expression in response to Hedgehog (Hh) signaling activation.1 GLI1 contains 5 conserved C2-H2 zinc finger domains that specifically bind DNA sequences in gene promoters to potentiate or repress the expression of target genes.2,3 Three structurally homologous family members—GLI1, GLI2, and GLI3—have been identified in mammalian cells; however, their biochemical properties and functions are highly variable.4,5 GLI2 and GLI3 have both C-terminal transcriptional activation and N-terminal repression domains. They are sequentially phosphorylated by multiple kinases (such as PKA, GSK3, and CK1) in their C-terminal regions, triggering proteolytic processing that converts the full-length forms (transcriptional activators) into truncated forms (transcriptional repressors).6 In contrast, GLI1 harbors only the C-terminal transcriptional activation domain and thus acts only as a transcriptional activator, providing key transcriptional output of Hh signaling.7
GLI1 is an oncogene implicated in the pathobiology of several neoplasms such as glioblastomas,8 basal cell carcinomas,9 medulloblastomas,10 and rhabdomyosarcomas.11 Previously, we demonstrated that the canonical Hh ligand-PTCH1-SMO-GLI1 axis is functional, and GLI1 is constitutively active, in a large subset of diffuse large B-cell lymphomas (DLBCL). We further demonstrated that the canonical Hh ligand-PTCH1-SMO-GLI1 axis plays important roles in cell proliferation, survival, and chemotolerance in this lymphoma subtype.12-16
Regulators of GLI1’s activities include SNF5, a core subunit of the adenosine trisphophate (ATP)-dependent SWItch/Sucrose Non-Fermentable chromatin remodeling complex, which modulates its transcriptional activities.17 Hh signaling, meanwhile, promotes the transcriptional activity of both GLI1 and GLI2 proteins by promoting their deacetylation via HDAC1 upregulation. Hh signaling pathway activity is inhibited by REN, an adaptor subunit of the Cullin-3–based ubiquitin ligase complex, which targets HDAC1 for ubiquitination and proteasome degradation.18 Activation of Hh signaling also strongly affects GLI1 protein stability.19-21 Little is known regarding the regulation of the GLI1’s transcriptional activities, despite its importance in both malignant and non-malignant biology.
The nuclear factor (NF-κB) pathway plays a critical role in B-cell physiology and contributes to the proliferation and survival of DLBCL cells.22 The IKK complex activates NF-κB via phosphorylation of the inhibitory molecule IkBα.23 Recent studies found the IKK complex also has key NF-κB–independent roles in a variety of physiologic and pathologic processes (eg, through the regulation of Myc and p73 transcription factors).24,25 In this report, we demonstrate for the first time that GLI1 also is a direct substrate of IKKβ. Together with other binding partners, IKKβ forms a multiprotein complex with GLI1 and regulates the stability of GLI1. This is important because elevated GLI1 protein levels (resulting from increased stability) dramatically accelerate tumor induction in mice.19
Methods
Cells and proliferation assays
Cells and cell culture procedures used in this study are described in the supplemental Material, available on the Blood Web site. For the coculture experiments, HS-5 cells were plated in 6-well plates using RPMI 1640 medium with 2% fetal bovine serum and allowed to attach and grow for 24 hours. Then trans-well cell cultures, with 0.4-μM pores, were inserted containing DLBCL cells. The coculture between HS-5 and DLBCL cells were maintained for 24 hours. Wherever they are mentioned, cells were treated with tumor necrosis factor-α (TNFα) (20 ng/mL) or TPCA1 (15 mg/kg) and/or GLI1 antagonist (GANT61; 40 mg/kg) for the indicated time periods. Transfections, lentiviral infections,12 and cell viability assays were performed as previously described.26
Constructs and RNA interference
Constructs and RNA interference assays used in this study are shown in the supplemental Material. Wild-type IKKα, IKKβ, and kinase-dead (K44A) constructs were described previously.27
Protein analysis and mass spectrometry
Cell lysis, immunoblotting, and immunoprecipitation assays were previously described.28,29 To detect protein ubiquitination, cells were lysed in RIPA buffer and boiled for 5 minutes at 95°C. Supernatants were diluted tenfold with normal 1% Triton lysis buffer, incubated with an antibody overnight at 4°C followed by incubation with 50% protein A/G ultralink resin slurry (Thermo Scientific) for 2 hours. Immobilized complexes were washed in RIPA lysis buffer, eluted, and subjected to immunoblotting. The antibodies used for immunoblotting are described in the supplemental Material.
The calf intestinal alkaline phosphatase (CIP) assay was performed as described.30 For mass spectrometric phosphopeptide analysis, 293T cells were transiently cotransfected with constructs carrying full-length GLI1 and wild-type IKKβ. GLI1 protein was resolved on sodium dodecyl sulfate-polyacrylamide gel electrophoresis by combining 5 GLI1 immunoprecipitation samples and visualized by Coomassie Blue staining. In-gel digestion analyses of immunoprecipitated samples were analyzed by Nano LC-MS/MS on an Orbitrap-XL mass spectrometer (Thermo Scientific).31
Proteins were identified by a database search of the fragment spectra against the National SwissProt protein database (EBI) using Mascot v.2.3 (Matrix Science) and Sequest (v.1.20) via Proteome Discoverer v.1.3 (Thermo Fisher Scientific). Phosphopeptide matches were analyzed using a combination of PhosphoRS implemented in Proteome Discoverer and manual interpretation.32
In vitro interaction and kinase assay
Glutathione S-transferase (GST) beads or beads bound to the indicated GST–IKKβ (Abcam) protein were equilibrated in phosphate-buffered saline at 4°C for 4 hours. Reactions using recombinant full-length purified GLI1 (isolated with anti-GLI1 IP) were incubated at 4°C for 4 hours. Beads were washed 3 times in phosphate-buffered saline, and bound protein was eluted and analyzed by immunoblotting.
400-1106 fragment of hGLI1 (GLI1F) or GLI1FTM was cloned into pGEX-6p1 construct and expressed in BL21 (DE3). Recombinant GST, GST-GLI1F, and GST-GLI1FTM proteins were isolated using Agarose glutathione (Sigma-Aldrich) and eluted with reduced glutathione. In vitro kinase assays were performed as described previously.33
GLI1 luciferase reporter assay and quantitative real-time PCR analysis
GLI1 luciferase reporter assay and quantitative real-time polymerase chain reaction (qRT-PCR) analyses were performed as described previously.14 Primers used for qRT-PCR analyses were obtained from Applied Biosystems.
Site-directed mutagenesis
Site-directed mutagenesis was performed as described previously.14 The sequences of primers used for mutagenesis are shown in the supplemental Material.
Statistical analysis
For isobologram graph analysis, cell viability measurements (ie, 50% growth inhibition) were determined by cell viability assays, as previously described.26 Data of individual doses (ie, of drug 1 and drug 2) were plotted on the x- and y-axis, respectively. The combination index (CI) of the Chou and Talalay method was used to determine the combination of drug synergy.34 Calcusyn software was used to calculate the CI.
Results
IKKβ is a novel binding partner of GLI1
To identify proteins that regulate GLI1 activity, we used liquid chromatography tandem mass spectrometry after immunoprecipitation of overexpressed GLI1. We identified that the catalytic subunit of the IKK complex, IKKβ, was one of the proteins associated with GLI1 (Figure 1A). To validate the association between endogenous GLI1 with IKKβ, lysates of 293T and SUDHL4 cells were immunoprecipitated with GLI1 antibody. IKKβ coimmunoprecipitated with GLI1 (Figure 1B), and the association between endogenous GLI1 and IKKβ was validated by reciprocal immunoprecipitation experiments (Figure 1C). In vitro assays with recombinant GST-IKKβ protein showed that IKKβ interacts with GLI1 (Figure 1D). Therefore, these data suggest that IKKβ and GLI1 are integral constituents of the same complex.
IKKβ associates with GLI1 transcription factor. (A) To identify GLI1-interacting partners, we cotransfected full-length GLI1 construct in 293T cells, and the GLI1 complex was purified using anti-GLI1 antibody. Peptides resulting from digestion of GLI1 with proteases were analyzed by mass spectrometry. Identified proteins are shown in color according to their biological function. Red color depicts cell-cell signaling; purple, green, and blue colors depict signal transduction, protein biogenesis, and cell cycle, respectively. Endogenous immunoprecipitation of (B) GLI1 or (C) IKKβ in 293T and SUDHL4 cells. (D) GST-tagged IKKβ and affinity-purified GLI1 were used for an in vitro pull-down assay. (E) IKKα and IKKβ constructs were coexpressed in 293T cells with vector or full-length flag-tagged (FL)-GLI1, and total cell lysates were subjected to anti-GLI1 immunoprecipitation. (F) FL-GLI1 construct was coexpressed with vector, IKKβ, or IKKβ-KD (K44A) in 293T cells, and total cell lysates were subjected to anti-GLI1 immunoprecipitation. (G) IKKβ or IKKβ-KD was coexpressed with vector or FL-GLI1 in 293T cells, and total cell lysates were subjected to anti-IKKβ immunoprecipitation. Phosphorylation of IKKβ (Ser-181) confirmed activation of IKKβ in IKKβ-transfected cells. Red arrows depict slow-migratory GLI1 protein bands.
IKKβ associates with GLI1 transcription factor. (A) To identify GLI1-interacting partners, we cotransfected full-length GLI1 construct in 293T cells, and the GLI1 complex was purified using anti-GLI1 antibody. Peptides resulting from digestion of GLI1 with proteases were analyzed by mass spectrometry. Identified proteins are shown in color according to their biological function. Red color depicts cell-cell signaling; purple, green, and blue colors depict signal transduction, protein biogenesis, and cell cycle, respectively. Endogenous immunoprecipitation of (B) GLI1 or (C) IKKβ in 293T and SUDHL4 cells. (D) GST-tagged IKKβ and affinity-purified GLI1 were used for an in vitro pull-down assay. (E) IKKα and IKKβ constructs were coexpressed in 293T cells with vector or full-length flag-tagged (FL)-GLI1, and total cell lysates were subjected to anti-GLI1 immunoprecipitation. (F) FL-GLI1 construct was coexpressed with vector, IKKβ, or IKKβ-KD (K44A) in 293T cells, and total cell lysates were subjected to anti-GLI1 immunoprecipitation. (G) IKKβ or IKKβ-KD was coexpressed with vector or FL-GLI1 in 293T cells, and total cell lysates were subjected to anti-IKKβ immunoprecipitation. Phosphorylation of IKKβ (Ser-181) confirmed activation of IKKβ in IKKβ-transfected cells. Red arrows depict slow-migratory GLI1 protein bands.
IKKβ is a catalytic subunit of the IKK complex, which contains an additional catalytic IKKα subunit. To examine whether GLI1 forms a complex with IKKα, we performed coimmunoprecipitation assays using 293T cells that were transiently cotransfected with constructs encoding full-length flag-tagged GLI1 (FL-GLI1) and IKKα or IKKβ. Although both subunits of the IKK complex were detected in the immunoprecipitated GLI1 protein complex, only constructs encoding GLI1 and IKKβ increased GLI1 protein levels and resulted in the presence of a slow migratory GLI1 band (Figure 1E). Consequently, we examined whether the kinase domain of IKKβ is critical for its association with GLI1. For this, 293T cells were transiently transfected with constructs carrying FL-GLI1 and wild-type IKKβ or kinase-dead IKKβ mutant (K44A). The immunoprecipitation assays showed that GLI1 associates with the wild-type IKKβ but not with the kinase-dead IKKβ mutant (Figure 1F-G), suggesting a functional role of IKKβ activity in the association and accumulation of GLI1.
IKKβ modulates GLI1 transcriptional activity
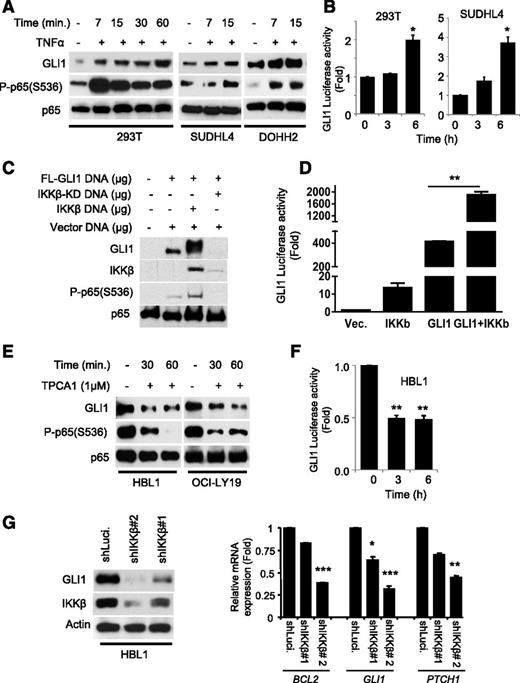
TNFα is known to be a potent activator of the NF-κB pathway through activation of IKKs’ kinase activity.23 To address the functional relevance of IKKβ in regulating the abundance of GLI1, we first performed immunoblot analysis of DLBCL and 293T cell lysates stimulated with TNFα. TNFα rapidly increased GLI1 protein levels (Figure 2A) and GLI1 transcriptional activity (Figure 2B) as measured by GLI1 luciferase reporter assays. Similar results were observed in 293T cells transiently transfected with constructs carrying GLI1 and IKKβ (Figure 2C-D), suggesting that the TNFα-mediated accumulation of GLI1 and its transcriptional output depend on IKKβ activity.
GLI1 transcriptional activity is modulated by IKKβ. (A) Immunoblot analysis of GLI1 in indicated cells stimulated with or without TNFα (20 μg/mL) for indicated time periods. Phosphorylation of the P65 (Ser-536) subunit of NF-κB confirmed IKKβ-mediated activation of the NF-κB pathway in the TNFα-stimulated cells. (B) 8X GLI1 luciferase and Renilla constructs were cotransfected in indicated cells for 48 hours and subjected to luciferase reporter assay. Results are normalized to Renilla luciferase and expressed as fold change in relative luciferase activity compared with control. Data represent the mean and standard deviation of 3 independent experiments (*P < .05). (C) Immunoblot analysis of GLI1 in 293T cells transiently transfected with constructs carrying FL-GLI1 and the IKKβ or IKKβ-KD. (D) GLI1 luciferase reporter analysis of 293T cells transiently transfected with constructs carrying vector, IKKβ, or FL-GLI1 and IKKβ. Results are normalized to Renilla luciferase and expressed as fold change in relative luciferase activity compared with control. Data represent the mean and standard deviation of 3 independent experiments (**P < .005). (E) Immunoblot analysis of GLI1 in DLBCL cells treated with or without TPCA1 for indicated time periods. (F) GLI1 luciferase reporter analysis of HBL1 cells treated with or without TPCA1. Results are normalized to Renilla luciferase and expressed as fold change in relative luciferase activity compared with control. Data represent the mean and standard deviation of 3 independent experiments (** P < .005). (G) HBL1 cells were transduced with lentiviral particles expressing shRNA-targeting luciferase (control) and IKKβ shRNAs. The transduced cells were selected with puromycin and subjected to immunoblotting. The same cells as described in (G) were used for qRT-PCR analysis of BCL2, GLI1, and PTCH1 mRNA expression. Results are normalized to GAPDH mRNA level and expressed as fold changes in mRNA expression compared with control. Data represent the mean and standard deviation of 2 independent experiments (*P < .05; ***P < .0005).
GLI1 transcriptional activity is modulated by IKKβ. (A) Immunoblot analysis of GLI1 in indicated cells stimulated with or without TNFα (20 μg/mL) for indicated time periods. Phosphorylation of the P65 (Ser-536) subunit of NF-κB confirmed IKKβ-mediated activation of the NF-κB pathway in the TNFα-stimulated cells. (B) 8X GLI1 luciferase and Renilla constructs were cotransfected in indicated cells for 48 hours and subjected to luciferase reporter assay. Results are normalized to Renilla luciferase and expressed as fold change in relative luciferase activity compared with control. Data represent the mean and standard deviation of 3 independent experiments (*P < .05). (C) Immunoblot analysis of GLI1 in 293T cells transiently transfected with constructs carrying FL-GLI1 and the IKKβ or IKKβ-KD. (D) GLI1 luciferase reporter analysis of 293T cells transiently transfected with constructs carrying vector, IKKβ, or FL-GLI1 and IKKβ. Results are normalized to Renilla luciferase and expressed as fold change in relative luciferase activity compared with control. Data represent the mean and standard deviation of 3 independent experiments (**P < .005). (E) Immunoblot analysis of GLI1 in DLBCL cells treated with or without TPCA1 for indicated time periods. (F) GLI1 luciferase reporter analysis of HBL1 cells treated with or without TPCA1. Results are normalized to Renilla luciferase and expressed as fold change in relative luciferase activity compared with control. Data represent the mean and standard deviation of 3 independent experiments (** P < .005). (G) HBL1 cells were transduced with lentiviral particles expressing shRNA-targeting luciferase (control) and IKKβ shRNAs. The transduced cells were selected with puromycin and subjected to immunoblotting. The same cells as described in (G) were used for qRT-PCR analysis of BCL2, GLI1, and PTCH1 mRNA expression. Results are normalized to GAPDH mRNA level and expressed as fold changes in mRNA expression compared with control. Data represent the mean and standard deviation of 2 independent experiments (*P < .05; ***P < .0005).
To examine whether inhibition of IKKβ decreases GLI1 protein levels and activity, we treated constitutive NF-κB active DLBCL cell lines (HBL1 and OCI-LY19) with an IKKβ inhibitor, TPCA1.35 As expected, TPCA1-treated DLBCL cells exhibited a decrease in GLI1 protein levels (Figure 2E and supplemental Figure 1) and a decrease in GLI1 transcriptional activity (Figure 2F). Concordantly, we observed decreased GLI1 levels (Figure 2G, left panel) and decreased expression of GLI1 direct-downstream genes (PTCH1, BCL2, and GLI1) in IKKβ-depleted DLBCL cell lines (Figure 2G, right panel, and supplemental Figure 2). Altogether, these data strongly support the functional importance of IKKβ in accumulation and transcriptional activity of GLI1.
IKKβ suppresses GLI1 ubiquitination
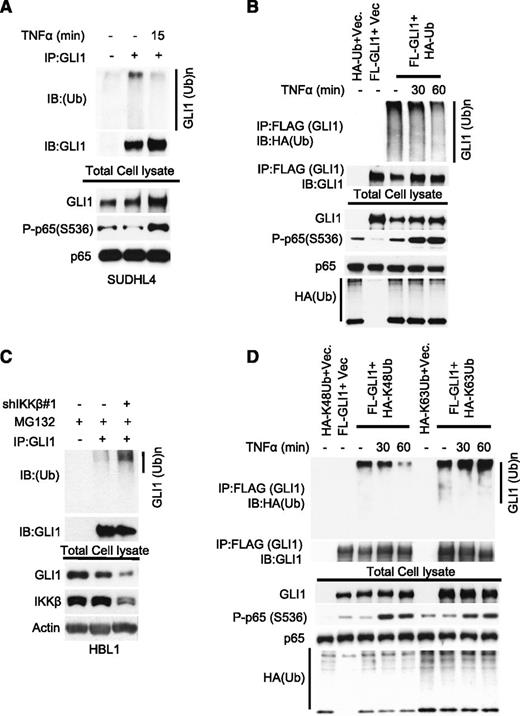
Given that short TNFα-induced activation of IKKβ rapidly increases the protein expression of GLI1, we investigated whether IKKβ participates in the regulation of GLI1 protein stability. We first analyzed total endogenous GLI1 ubiquitination in SUDHL4 cells in the presence or absence of TNFα. TNFα stimulation decreased total GLI1 ubiquitination in SUDHL4 (Figure 3A) and in 293T cells transiently transfected with GLI1 and HA-ubiquitin constructs (Figure 3B), without significantly changing expression of known direct GLI1 target genes (supplemental Figure 3). Next, we analyzed endogenous GLI1 ubiquitination in IKKβ-depleted HBL1 cells in the presence or absence of the proteasome inhibitor MG132. As expected, we observed an increase in total GLI1 ubiquitination in IKKβ-depleted HBL1 cells (Figure 3C). These observations suggest that IKKβ is involved in regulating GLI1 protein ubiquitination. Because K48-linked ubiquitination is known to regulate protein degradation,36 we next tested whether TNFα-induced IKKβ activation affects K48 or K63-linked GLI1 ubiquitination. TNFα-triggered IKKβ activation decreased K48-linked, but not K63-linked GLI1 ubiquitination (Figure 3D), implicating IKKβ activity in the stabilization of GLI1 by reducing K48-linked GLI1 ubiquitination.
TNFα-mediated activation of IKKβ suppresses GLI1 ubiquitination. (A) Immunoprecipitation analysis of ubiquitinated-GLI1 in SUDHL4 cells stimulated with or without TNFα (20 μg/mL) for 15 minutes. (B) Indicated constructs were coexpressed in 293T cells. Before cell lysis, cells were stimulated with or without TNFα for indicated time periods. Total cell lysates were subjected to anti-GLI1 immunoprecipitation. Bound ubiquitinated-GLI1 was detected by anti-HA antibody. (C) Immunoprecipitation analysis of total ubiquitinated-GLI1 in control (Cont.) or partially IKKβ-depleted HBL1 cells after treated with proteasome inhibitor MG132 (10 μg/mL) for 6 hours. (D) Immunoprecipitation analysis of K48 or K63-linked ubiquitinated-GLI1 in 293T cells transfected with FL-GLI1 and HA-K48 or HA-K63–linked ubiquitin constructs. Before cell lysis, cells were stimulated with or without TNFα (20 μg/mL) for indicated time periods.
TNFα-mediated activation of IKKβ suppresses GLI1 ubiquitination. (A) Immunoprecipitation analysis of ubiquitinated-GLI1 in SUDHL4 cells stimulated with or without TNFα (20 μg/mL) for 15 minutes. (B) Indicated constructs were coexpressed in 293T cells. Before cell lysis, cells were stimulated with or without TNFα for indicated time periods. Total cell lysates were subjected to anti-GLI1 immunoprecipitation. Bound ubiquitinated-GLI1 was detected by anti-HA antibody. (C) Immunoprecipitation analysis of total ubiquitinated-GLI1 in control (Cont.) or partially IKKβ-depleted HBL1 cells after treated with proteasome inhibitor MG132 (10 μg/mL) for 6 hours. (D) Immunoprecipitation analysis of K48 or K63-linked ubiquitinated-GLI1 in 293T cells transfected with FL-GLI1 and HA-K48 or HA-K63–linked ubiquitin constructs. Before cell lysis, cells were stimulated with or without TNFα (20 μg/mL) for indicated time periods.
IKKβ activity is essential for ITCH-mediated GLI1 ubiquitination
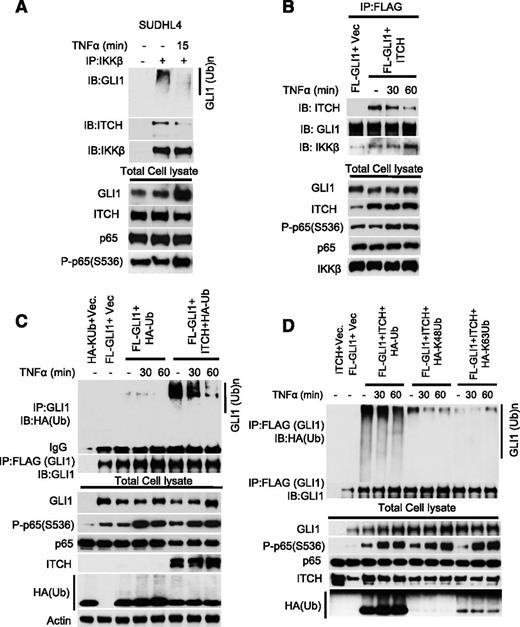
ITCH is an E3 ligase that ubiquitinates and targets GLI1 for proteosomal degradation.19-21,37 The mechanism involved in regulating ITCH-mediated GLI1 ubiquitination remains unknown. To examine whether IKKβ activity regulates the dynamic binding between ITCH and GLI1, lysates of SUDHL4 cells stimulated with TNFα were subjected to IKKβ immunoprecipitation. ITCH and GLI1 proteins coimmunoprecipitated with IKKβ (Figure 4A), and TNFα stimulation decreased the binding between ITCH and GLI1 (Figure 4A). The association between ITCH and IKKβ was further validated by reciprocal anti-GLI1 immunoprecipitation experiments in 293T cells transiently transfected with GLI1 and ITCH constructs (Figure 4B). We also assessed the level of GLI1 ubiquitination in 293T cells transiently transfected with ITCH and GLI1 constructs after TNFα stimulation. Transient transfection of ITCH increased total and K48-linked ubiquitination of GLI1 that was decreased by TNFα (Figure 4C-D). Our data suggest that TNFα-mediated IKKβ activity regulates the binding between ITCH and GLI1 and therefore the ITCH-mediated GLI1 ubiquitination.
GLI1 ubiquitination requires IKKβ and the E3 ligase ITCH. (A) SUDHL4 cells were stimulated with or without TNFα (20 μg/mL) for 15 minutes. Total cell lysates were subjected to anti-GLI1 immunoprecipitation. Bound proteins were detected with indicated antibodies. (B) FL-GLI1 was coexpressed in 293T cells in the presence or absence of ITCH construct. After 48 hours, cells were stimulated with or without TNFα for indicated time periods. Total cell lysates were subjected to anti-GLI1 immunoprecipitation, and bound proteins were detected with indicated antibodies. (C) Immunoprecipitation analysis of ubiquitinated-GLI1 in TNFα-stimulated 293T cells transfected with indicated constructs. (D) Immunoprecipitation analysis of K48 or K63 ubiquitinated-GLI1 in TNFα-stimulated 293T cells transfected with indicated constructs.
GLI1 ubiquitination requires IKKβ and the E3 ligase ITCH. (A) SUDHL4 cells were stimulated with or without TNFα (20 μg/mL) for 15 minutes. Total cell lysates were subjected to anti-GLI1 immunoprecipitation. Bound proteins were detected with indicated antibodies. (B) FL-GLI1 was coexpressed in 293T cells in the presence or absence of ITCH construct. After 48 hours, cells were stimulated with or without TNFα for indicated time periods. Total cell lysates were subjected to anti-GLI1 immunoprecipitation, and bound proteins were detected with indicated antibodies. (C) Immunoprecipitation analysis of ubiquitinated-GLI1 in TNFα-stimulated 293T cells transfected with indicated constructs. (D) Immunoprecipitation analysis of K48 or K63 ubiquitinated-GLI1 in TNFα-stimulated 293T cells transfected with indicated constructs.
IKKβ phosphorylates GLI1
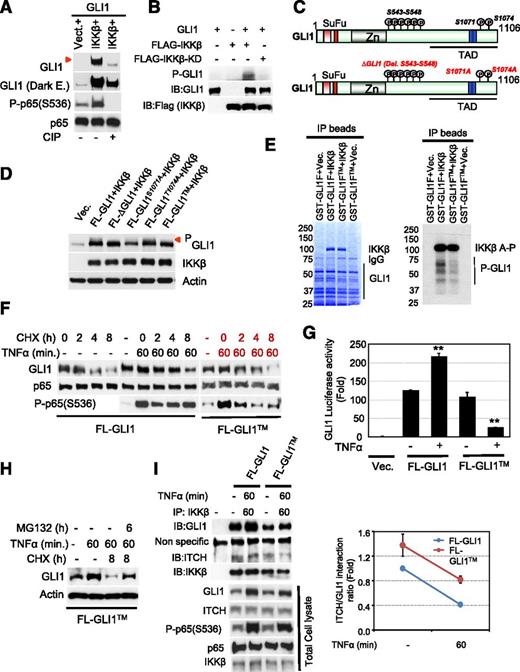
We initially observed that cotransfection of GLI1 with IKKβ resulted in the appearance of a slow-migratory GLI1 band that was not detected in cells transfected with kinase-dead IKKβ mutant (Figures 1F-G). IKKβ is a serine-threonine protein kinase that activates the NF-κB pathway by phosphorylating IκB.38 For this reason, we were interested to determine whether the detected slow-migratory GLI1 band is phosphorylated GLI1. 293T cell lysates collected after transfection of full-length GLI1 and IKKβ constructs were incubated with and without CIP. As is shown in Figure 5A, incubation with CIP (lane 3) resulted in loss of the slow-migratory GLI1 band supporting that this band represents phosphorylated GLI1. Furthermore, we performed an in vitro kinase assay in 293T cells transfected with epitope-tagged expression vectors containing wild-type or kinase-dead IKKβ. Immunoprecipitated kinase was assayed with affinity-purified GLI1 as a substrate in the presence of (γ32P) ATP and confirmed that full-length purified GLI1 was phosphorylated by IKKβ (Figure 5B).
GLI1 is a bona fide substrate of IKKβ. (A) 293T cells were transiently cotransfected with constructs carrying GLI1 and IKKβ wild-type. Cell lysates were incubated with or without calf intestinal alkaline phosphatase (CIP) and subjected to immunoblotting. The red arrow indicates phosphorylated GLI1 protein band. (B) Epitope-tagged expression vectors IKKβ or IKKβ-KD constructs were transfected into 293T cells. Immunoprecipitated kinase was assayed with affinity-purified GLI1 as a substrate in the presence of (γ32P) ATP. (C) To identify IKKβ-dependent GLI1 phosphorylation sites, we cotransfected FL-GLI1 and IKKβ constructs in 293T cells, and the GLI1-complex was purified using anti-GLI1 antibody. Purified samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and band representing GLI protein was cut and analyzed by nanospray ion trap mass spectrometry. Eight phosphorylation sites were identified in the C-terminal region of GLI1 (S543-548, S1071, and T1074) as shown in the schematic diagram. (D) Immunoblot analysis of phosphorylated GLI1 in 293T cells transiently transfected with indicated GLI1 mutant constructs and IKKβ. (E) GST-purified GLI1 fragment (GLI1F; 411aa-1106aa) or triple-mutant GLI1 fragment (GLI1FTM) protein was incubated with immunoprecipitated IKKβ kinase in the presence of (γ32P) ATP and analyzed by in vitro kinase assay. (F) Cycloheximide (CHX) chase analysis of GLI1 or indicated full-length GLI1TM construct was expressed in 293T cells in the presence or absence of CHX (60 μg/mL) for indicated time periods. After TNFα (20 μg/mL) stimulation, cell lysates from each time point were immunoblotted with anti-GLI1, anti-P-p65 (Ser536), and anti-p65 antibodies. (G) GLI1 luciferase reporter analysis of 293T cells transiently transfected with indicated constructs with or without TNFα stimulation for 6 hours. Results are normalized to Renilla luciferase and expressed as fold change in relative luciferase activity compared with control. Data represent the mean and standard deviation of 2 independent experiments (**P < .005). (H) FL-GLI1TM construct was transiently transfected in 293T cells. After 48 hours, cells were stimulated with TNFα (20 μg/mL) for 60 minutes after CHX (60 μg/mL) and MG132 (30 μg/mL) treatment of indicated time periods. Cell lysates from each time point were immunoblotted with anti-GLI1 antibody. (I) 293T cells were cotransfected with full-length GLI1 and GLI1TM mutant construct, and ITCH-IKKβ-GLI1 complex was purified using anti-IKKβ antibody and subjected to immunoblotting. Each component of GLI1 complex immunoblots were analyzed by densitometry, normalized with IKKβ, and plotted to determine normalized ITCH/GLI1 association.
GLI1 is a bona fide substrate of IKKβ. (A) 293T cells were transiently cotransfected with constructs carrying GLI1 and IKKβ wild-type. Cell lysates were incubated with or without calf intestinal alkaline phosphatase (CIP) and subjected to immunoblotting. The red arrow indicates phosphorylated GLI1 protein band. (B) Epitope-tagged expression vectors IKKβ or IKKβ-KD constructs were transfected into 293T cells. Immunoprecipitated kinase was assayed with affinity-purified GLI1 as a substrate in the presence of (γ32P) ATP. (C) To identify IKKβ-dependent GLI1 phosphorylation sites, we cotransfected FL-GLI1 and IKKβ constructs in 293T cells, and the GLI1-complex was purified using anti-GLI1 antibody. Purified samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and band representing GLI protein was cut and analyzed by nanospray ion trap mass spectrometry. Eight phosphorylation sites were identified in the C-terminal region of GLI1 (S543-548, S1071, and T1074) as shown in the schematic diagram. (D) Immunoblot analysis of phosphorylated GLI1 in 293T cells transiently transfected with indicated GLI1 mutant constructs and IKKβ. (E) GST-purified GLI1 fragment (GLI1F; 411aa-1106aa) or triple-mutant GLI1 fragment (GLI1FTM) protein was incubated with immunoprecipitated IKKβ kinase in the presence of (γ32P) ATP and analyzed by in vitro kinase assay. (F) Cycloheximide (CHX) chase analysis of GLI1 or indicated full-length GLI1TM construct was expressed in 293T cells in the presence or absence of CHX (60 μg/mL) for indicated time periods. After TNFα (20 μg/mL) stimulation, cell lysates from each time point were immunoblotted with anti-GLI1, anti-P-p65 (Ser536), and anti-p65 antibodies. (G) GLI1 luciferase reporter analysis of 293T cells transiently transfected with indicated constructs with or without TNFα stimulation for 6 hours. Results are normalized to Renilla luciferase and expressed as fold change in relative luciferase activity compared with control. Data represent the mean and standard deviation of 2 independent experiments (**P < .005). (H) FL-GLI1TM construct was transiently transfected in 293T cells. After 48 hours, cells were stimulated with TNFα (20 μg/mL) for 60 minutes after CHX (60 μg/mL) and MG132 (30 μg/mL) treatment of indicated time periods. Cell lysates from each time point were immunoblotted with anti-GLI1 antibody. (I) 293T cells were cotransfected with full-length GLI1 and GLI1TM mutant construct, and ITCH-IKKβ-GLI1 complex was purified using anti-IKKβ antibody and subjected to immunoblotting. Each component of GLI1 complex immunoblots were analyzed by densitometry, normalized with IKKβ, and plotted to determine normalized ITCH/GLI1 association.
To identify IKKβ-dependent GLI1 phosphorylation sites, we performed mass spectrometry phosphopeptide analysis as described in Methods. Nanospray ion trap mass spectrometry of peptides digested from the immunoprecipitated IKKβ-GLI1 complex identified 8 evolutionarily conserved phosphorylation sites (supplemental Figure 4) in the C-terminal region of GLI1 (Figure 5C and supplemental Table 1). We then generated a truncated GST-GLI1 construct (GLI1F; 401-1106aa) harboring all the probable IKKβ-dependent GLI1 phosphorylation sites (identified by mass spectrometry) and performed in vitro kinase assays. Although GST-purified, truncated GLI1F protein was highly unstable (supplemental Figure 5), the in vitro kinase assay showed that the GST-GLI1F was extensively phosphorylated by wild-type but not by kinase-dead IKKβ (supplemental Figure 6). Altogether, these data strongly suggest that GLI1 is phosphorylated by IKKβ.
Characterization of the IKKβ-mediated GLI1 phosphorylation sites
The C-terminal region of GLI1 harbors phosphorylated serine-proline residues, a consensus sequence previously shown to participate in ITCH-induced ubiquitination and degradation of GLI1.39 We hypothesized that IKKβ-mediated phosphorylation of GLI1 may inhibit ITCH-mediated GLI1 ubiquitination, contributing to GLI1 protein stabilization. To address this possibility, we individually examined the phosphorylation status of GLI1 after deleting a cluster of 6 serine amino acid residues (S543-S548; ΔGLI1) or replacing serine 1071 (S1071A) or threonine 1074 (T1074A) with alanine from the full-length GLI1 construct using anti-GLI1 antibody (that recognizes phosphorylated GLI1 protein). Although we observed a varying degree of inhibition in GLI1 phosphorylation in GLI1-mutant constructs (Figure 5D), cells cotransfected with the ΔGLI1 or S1071A mutant and IKKβ constructs exhibited marked decrease in the slow-migratory GLI1 phosphorylation band compared with the cells cotransfected with full-length GLI1 and IKKβ (Figure 5D). Similarly, we observed rapid degradation of GLI1 in the cycloheximide (CHX)-treated 293T cells transiently transfected with ΔGLI1 or S1071A mutants after stimulation with TNFα, suggesting that the cluster of these 6 serine amino acid residues and S1071 are phosphorylation sites that contribute to GLI1 protein stability more than T1074 (supplemental Figure 7A-B).
We then created a triple-mutant GST-GLI1FTM (ΔGLI1+S1071A+T1074A) construct and performed in vitro kinase assays. These assays confirmed that the truncated wild-type GST-GLI1F, but not the GST-GLI1FTM, was phosphorylated by IKKβ (Figure 5E). Although 8 evolutionarily conserved GLI1 phosphorylation sites were confirmed using in vitro kinase assays, we were still able to detect weak GLI1 phosphorylation in the GST-GLI1FTM construct (Figure 5E). These data suggest that additional phosphorylation site(s) may exist. Similarly, cell-expressing GLI1FTM construct rapidly degraded GLI1 (Figure 5F) in a proteasome-dependent manner compared with cells expressing wild-type GLI1 or other constructs after TNFα stimulation (Figure 5H). Concordantly, cells expressing the GLI1FTM construct have decreased GLI1 transcriptional activity in the presence of TNFα (Figure 5G) supporting the role of these sites in GLI1 transcriptional activity.
To study further the functionality of these GLI1 sites in GLI1 ubiquitination and degradation, we explored whether removal or mutation of these sites (GLI1TM mutant) interferes with the dynamic interaction between IKKβ, GLI1, and ITCH. We observed decreased association between GLI1 with IKKβ in the GLI1TM (Figure 5I), suggesting that these GLI1 sites are important for the interaction with IKKβ. Moreover, we observed increased interaction of ITCH with GLI1 in TNFα-stimulated 293T cells transiently transfected with GLI1TM compared with wild-type GLI1 (Figure 5I). Altogether, these data provide evidence that IKKβ by phosphorylating GLI1 at multiple sites regulates ITCH-mediated GLI1 degradation.
High NF-κB activity is associated with high GLI1 nuclear expression
Activation of the NF-κB signaling pathway is a hallmark of different types of lymphomas, especially DLBCL.22 We hypothesized that DLBCL with high constitutive NF-κB activity (as measured by detection of nuclear p65) must exhibit high GLI1 nuclear expression. As expected, nuclear expression of p65 significantly correlated with GLI1 nuclear expression in GCB (n = 16) and ABC (n = 15) subtypes of DLBCL (Figure 6A-B). These data further support the crosstalk between canonical NF-κB signaling and GLI1 in tumor specimens.
Clinical correlation between activated NF-κB and noncanonical Hh signaling pathways. (A) Immunofluorescence analysis of p65 and GLI1 in non-neoplastic lymph nodes and DLBCL tumors. Non-neoplastic lymph nodes (controls) and DLBCL tissue sections were stained with GLI1 (red) and p65 (green) antibodies. (B) The diagram shows the correlation between GLI1 and p65-integrated nuclear density expression (percentage) in GCB (n = 16) and ABC (n = 15) DLBCL subtypes.
Clinical correlation between activated NF-κB and noncanonical Hh signaling pathways. (A) Immunofluorescence analysis of p65 and GLI1 in non-neoplastic lymph nodes and DLBCL tumors. Non-neoplastic lymph nodes (controls) and DLBCL tissue sections were stained with GLI1 (red) and p65 (green) antibodies. (B) The diagram shows the correlation between GLI1 and p65-integrated nuclear density expression (percentage) in GCB (n = 16) and ABC (n = 15) DLBCL subtypes.
Inhibiting IKKβ and GLI1 has a synergistic effect on cell viability of DLBCL cells
Previously, we demonstrated that the canonical Hh signaling is functional and contributes to cell proliferation, survival, and chemotolerance in DLBCL.15,16 Herein, we report that IKKβ regulates the degradation and transcriptional activation of GLI1. This is of potential clinical interest because NF-κB signaling is frequently activated by mutations and/or via extracellular signals from the lymphoma microenvironment and plays a key role in DLBCL biology.
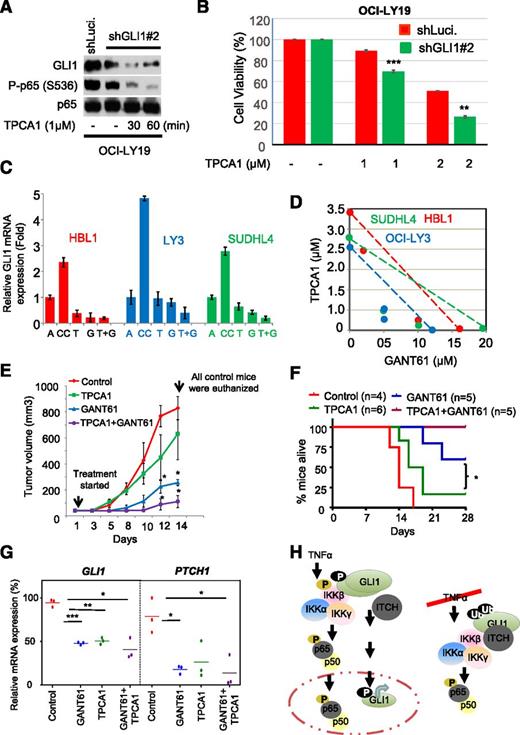
GLI1 is constitutively active in a large subset of DLBCL,13 and this activation may be mediated by canonical and noncanonical Hh signaling (eg, IKKβ as shown here). Therefore, we speculated that concomitant inhibition of GLI1 and NF-κB might further decrease DLBCL cell viability. To test this hypothesis, we knocked down GLI1 and studied the effect of inhibiting IKKβ activity on cell viability using TPCA1. TPCA1-treated GLI1 knockdown OCI-LY19 cells have lower GLI1 levels (Figure 7A) and cell viability than TPCA1-treated control cells (Figure 7B). Similarly, silencing of IKKβ decreased GLI1 protein levels and cell viability in control cells, whereas silencing of IKKβ in stable GLI1 knockdown cells further reduced DLBCL cell viability (supplemental Figure 8A-B).
Effects of combining inhibition of IKKβ with GLI1 on viability. OCI-LY19 cells were transduced with lentiviral particles expressing shRNA-targeting control (Luci.) and GLI1 (shGLI1#2). Luci. or stable GLI knockdown cells were treated with TPCA1 for indicated time periods. After 24 hours, cells were subjected to (A) immunoblotting and (B) cell viability assays. (C) Coculture of DLBCL cell lines with stromal cells (HS-5) in trans-well experiments. Cells were treated with or without TPCA1 and GANT61 or a combination of both for 24 hours. Cell lysates were then subjected to qRT-PCR analysis. Results are normalized to GAPDH mRNA level and expressed as fold changes in mRNA expression compared with control. Data represent the mean and standard deviation of 3 independent experiments. A, DLBCL cells alone; CC, DLBCL cells cocultured with HS-5 cells; G, GANT61; T, TPCA1; T+G, TPCA1+GANT61. (D) For isobologram graph analysis, DLBCL cells were cocultured with stromal cells (HS-5), treated with or without TPCA1 and GANT61 for 48 hours, and subjected to MTS assays. (E) NOD/SCID mice were inoculated subcutaneously in the flank with 5 × 106 OCI-LY10 cells. When the tumor was palpable, the mice were randomly subdivided into 4 groups (control, TPCA1, GANT61, and combined TPCA1 + GANT61), and the average tumor size for each group was determined. The value for each group was set to 0% (Day 0), and all subsequent changes in tumor size for each group will be expressed as a percentage change compared with the starting tumor mass. Control, GANT61, TPCA1, or combined (TPCA1 + GANT61) regimens were administered subcutaneously several centimeters away from the tumor site 3 times per week. Tumor sizes was assessed by standard calipers, and tumor volumes were determined weekly. Data represent the mean and standard deviation of 2 independent experiments (*P < .05). (F) Kaplan-Meier survival curves of mice treated with control, GANT61, TPCA1, or combined (TPCA1 + GANT61) regimens. The log-rank (Mantel Cox) test was used in survival curve analysis (*P < .05). (G) Proposed working model for noncanonical activation of GLI1 in response to TNFα-mediated activation of IKKβ.
Effects of combining inhibition of IKKβ with GLI1 on viability. OCI-LY19 cells were transduced with lentiviral particles expressing shRNA-targeting control (Luci.) and GLI1 (shGLI1#2). Luci. or stable GLI knockdown cells were treated with TPCA1 for indicated time periods. After 24 hours, cells were subjected to (A) immunoblotting and (B) cell viability assays. (C) Coculture of DLBCL cell lines with stromal cells (HS-5) in trans-well experiments. Cells were treated with or without TPCA1 and GANT61 or a combination of both for 24 hours. Cell lysates were then subjected to qRT-PCR analysis. Results are normalized to GAPDH mRNA level and expressed as fold changes in mRNA expression compared with control. Data represent the mean and standard deviation of 3 independent experiments. A, DLBCL cells alone; CC, DLBCL cells cocultured with HS-5 cells; G, GANT61; T, TPCA1; T+G, TPCA1+GANT61. (D) For isobologram graph analysis, DLBCL cells were cocultured with stromal cells (HS-5), treated with or without TPCA1 and GANT61 for 48 hours, and subjected to MTS assays. (E) NOD/SCID mice were inoculated subcutaneously in the flank with 5 × 106 OCI-LY10 cells. When the tumor was palpable, the mice were randomly subdivided into 4 groups (control, TPCA1, GANT61, and combined TPCA1 + GANT61), and the average tumor size for each group was determined. The value for each group was set to 0% (Day 0), and all subsequent changes in tumor size for each group will be expressed as a percentage change compared with the starting tumor mass. Control, GANT61, TPCA1, or combined (TPCA1 + GANT61) regimens were administered subcutaneously several centimeters away from the tumor site 3 times per week. Tumor sizes was assessed by standard calipers, and tumor volumes were determined weekly. Data represent the mean and standard deviation of 2 independent experiments (*P < .05). (F) Kaplan-Meier survival curves of mice treated with control, GANT61, TPCA1, or combined (TPCA1 + GANT61) regimens. The log-rank (Mantel Cox) test was used in survival curve analysis (*P < .05). (G) Proposed working model for noncanonical activation of GLI1 in response to TNFα-mediated activation of IKKβ.
We also investigated the effect of GLI1 and IKKβ inhibitors on DLBCL cells grown in the presence of HS-5 cells (stromal cells that are part of the reticular network of hematopoietic organs). The presence of HS-5 cells helps to recapitulate, to some extent, the prosurvival effect of the stromal microenvironment on the lymphoma cells.14,40 As previously described, coculturing stromal cells with DLBCL cells increased GLI1 expression.14 Cell viability after treatments with TPCA1 (IC50 = 1.0-3.5 μM) or GANT61 (IC50 = 10-20 μM) alone or in combination with TPCA1 was evaluated in DLBCL cells. Although each compound caused modest decrease in DLBCL cell viability, simultaneous treatment with GANT61 and TPCA1 led to a significant and synergistic decrease in cell viability in a panel of DLBCL cell lines (Figure 7C-D, supplemental Figure 9, and supplemental Table 2). Finally, we evaluated the synergistic effect of GANT61 and TPCA1 in a lymphoma xenograft model. Treatments with TPCA1 or GANT61 significantly decreased the expression of GLI1 target genes (PTCH1, BCL-2, and GLI1) (Figure 7G), resulting in reduced tumor growth (Figure 7E). Moreover, combined TPCA1-GANT61 treatment further reduced expression of GLI1 target genes and inhibited tumor growth at all time points compared with the control group, resulting in significantly increased survival (Figure 7E-G). Together, these findings provide a rationale for simultaneous targeting of both pathways in DLBCL.
Discussion
A number of transcription factors have been shown to be constitutively active in specific malignant neoplasms. This is of potential clinical significance, because they can be targets for therapeutic intervention.41 One of these transcription factors is GLI1, which is constitutively active in several cancers.8,10,13,16 We previously showed that GLI1 is constitutively activated in a large subset of DLBCL tumors and that GLI1 contributes to cell proliferation and survival of this lymphoma subtype.12,14,16 Here, we demonstrate that IKKβ binds GLI1 as part of a multiprotein complex and directly phosphorylates it, promoting stabilization of its intracellular levels. Such stabilization of GLI1 is known to promote the malignant phenotype in multiple cancers, and elevated GLI1 protein accelerates tumor induction in transgenic mice.19
Although IKKβ has been defined as a critical regulator of canonical NF-κB signaling activity in response to cytokines,38 this classic viewpoint has been expanded with the identification of novel roles of IKKs in the phosphorylation and stabilization of several transcription factors, such as Myc and p73.24,25 In accordance with these novel and NF-κB–independent functions of IKKβ,24,25 we show that IKKβ activity also plays a key role in the stabilization of GLI1 by interfering with its proteasomal degradation and results in increased GLI1 transcriptional activity in response to TNFα. Therefore, we show a key link in the signaling cascade that converts a signal emanating from the tumor microenvironment via TNFα into increased GLI1 stabilization and transcriptional activity through activation of IKKβ.
Ubiquitination-mediated degradation of GLI1 is a highly regulated process that requires specific E3 ubiquitin ligases.19-21 Recent studies suggest that at least 3 different E3 ligases may ubiquitinate and degrade GLI1: (1) SCFβ-Trcp1–based ubiquitination of GLI1, which is in part associated with activation of Hh signaling19 ; (2) Cullin3/HIB-SPOP–based ubiquitination and degradation of GLI120 ; and (3) ITCH-dependent degradation of GLI1.39 ITCH is a member of the HECT (homologous to E6-associated protein C terminus) family of E3 ubiquitin ligases and drives turnover of a number of transcription factors.39,42 Our data suggest that IKKβ is an essential component of the ITCH and GLI1 multiprotein ubiquitin-conjugating complex, and IKKβ-mediated phosphorylation of GLI1 within its C-terminal region is a regulatory step that inhibits GLI1 ubiquitination and degradation. We propose that phosphorylation of GLI1 by IKKβ sets in motion a sequence of events restricting the binding of GLI1 to ITCH, resulting in decreased ubiquitination and degradation. In the absence of TNFα, IKKβ activity is suppressed and GLI1 phosphorylation is restricted. In this state, unphosphophorylated GLI1 is more accessible to ITCH, resulting in its degradation (Figure 7G). Additional studies are required to understand the precise role of IKKβ (and additional factors such as deubiquitinases) in GLI1 stabilization.
In DLBCL, we previously demonstrated that canonical Hh ligand–mediated activation of GLI1 is functional and contributes to cell proliferation and survival.15,16,43 Here, we provide evidence for the presence of noncanonical GLI1 transcriptional activation mediated by IKKβ. Using in vitro stromal systems and in vivo models of DLBCL, we found that inhibiting IKKβ-mediated NF-κB activation and GLI1 synergistically decreased DLBCL cell viability, supporting a novel therapeutic approach for DLBCL based on concomitant inhibition of NF-κB and GLI1. This promising therapeutic strategy may also be useful on other types of tumors that have canonical NF-κB and GLI1 activation, as recently shown in a model of breast cancer.44
In summary, our findings suggest that GLI1 is a bona fide substrate of IKKβ and that IKKβ forms a multiprotein complex with GLI1 to regulate the stability of GLI1 and its transcriptional activity. This finding is biologically relevant because it links inflammatory signals from the tumor microenvironment with GLI1 transcriptional activity and tumor progression. Combined inhibition of IKKβ and GLI1 activities may represent a novel therapeutic strategy to treat DLBCL and other neoplasms with activated NF-κB and Hh signaling pathways.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Maria-Magdalena Georgescu, Dr Peter Zaphiropoulos, Dr Hui-Kuan Lin, and Noula Dattu Shembade for providing constructs; and Dr Stephen D. Nimer, Dr Arthur Zelent, and Dr Jonathan Schatz for critically reviewing the manuscript.
This work was supported by funds from University of Miami, CTSI Pilot Award Program 1UL1TR000460 (N.K.A., F.V.); The Translational Grant of The Leukemia & Lymphoma Society (F.V.); National Institutes of Health K08 Physician-Scientist Award 1 K08 CA143151-01 (F.V.); and the Dwoskin, Recio, and Rizzo family foundations (I.S.L.).
Authorship
Contribution: N.K.A. designed, performed experiments, analyzed the data, prepared figures, and wrote manuscript; C.H.K., G.A.K., and K.K. performed experiments; H.K. designed GLIF construct and in vitro kinase assay; V.T.M. performed immunofluorescence analysis; N.K.A., Y.T., C.H.K., K.K., and G.A.K. performed mice experiments; D.K. performed statistics; I.S.L., R.E.V., M.B., G.N.B., and F.V. supervised and critically analyzed data; and all authors commented on the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Nitin Kumar Agarwal, Division of Hematopathology, Department of Pathology, University of Miami, Sylvester Comprehensive Cancer Center, Miami, FL 33136; e-mail: n.agarwal1@med.miami.edu; and Francisco Vega, Division of Hematopathology, Department of Pathology, University of Miami, Sylvester Comprehensive Cancer Center, Miami, FL 33136; e-mail: fvega@med.miami.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal