In this issue of Blood, Lewinsohn and colleagues report on the inherited predisposition to hematologic malignancies (HMs) in 9 pedigrees with germ line mutations in the DEAD/H-box RNA helicase gene, DDX41.1
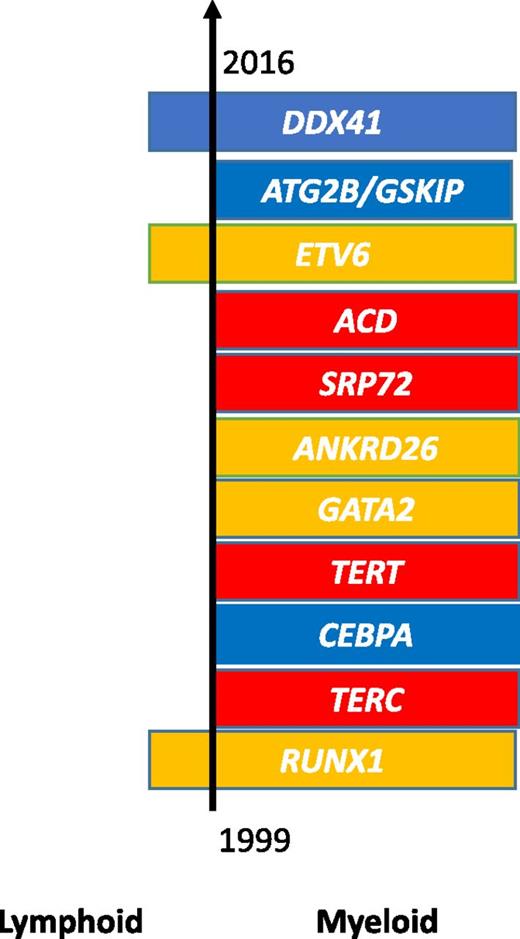
The genetic landscape of inherited HM. The order of the 11 established germ line mutations is depicted based on their date of discovery. Mutations are broadly assigned to 3 groups according to phenotype: HMs alone (blue), associated bone marrow failure syndromes (red), or characteristic cytopenias and/or platelet dysfunction (yellow). The incidence of these mutations varies considerably with >10 pedigrees reported for mutations in RUNX1, TERC, CEBPA, TERT, ANKRD26, GATA2, and now DDX41 with, in certain families/genes, apparent clustering of myeloid and lymphoid malignancies.
The genetic landscape of inherited HM. The order of the 11 established germ line mutations is depicted based on their date of discovery. Mutations are broadly assigned to 3 groups according to phenotype: HMs alone (blue), associated bone marrow failure syndromes (red), or characteristic cytopenias and/or platelet dysfunction (yellow). The incidence of these mutations varies considerably with >10 pedigrees reported for mutations in RUNX1, TERC, CEBPA, TERT, ANKRD26, GATA2, and now DDX41 with, in certain families/genes, apparent clustering of myeloid and lymphoid malignancies.
The current study expands on the first report of these mutations by Polprasert and colleagues,2 by recognizing further clinical and molecular heterogeneity in this genetic subgroup, with a newly identified predisposition to lymphoproliferative neoplasms and the discovery of germ line missense mutations in over 40% of families.
Next-generation sequencing (NGS) approaches are helping to elucidate the genomic landscape of inherited HMs at a remarkable pace. The recognition of inherited forms of disease can be challenging as patients themselves may be unaware of their predisposition coupled with a wide variation in the age of onset and disease phenotype. To date, research has broadly assembled leukemia predisposition syndromes into 3 groups characterized by HMs alone (CEBPA,3 ATG2B/GSKIP,4 and, most recently, DDX412 ), associated bone marrow failure syndromes (TERC,5 TERT,6 SRP72,5 ACD7 ), and HMs with preceding cytopenias and/or platelet dysfunction (RUNX1,8 GATA2,5 ANKRD26,5 ETV69,10 ) (see figure).
Lewinsohn and colleagues1 report on the inherited predisposition to HMs with germ line mutations in DDX41. This collaborative study identified 300 families with evidence of inherited HM, offering an unrivaled opportunity to identify new susceptibility loci and to capture the phenotypic and genetic diversity within a given genetic subgroup. In light of the cumulative report of 16 pedigrees with germ line mutations,1,2 DDX41 now represents a significant new addition to the genetic landscape of inherited HM.
In the current series, the authors report 3 additional families with the recently reported p.D140GfsX2 frameshift mutation, all associated with late-onset acute myeloid leukemia (AML). Other pedigrees appear distinctive, and include patients with myelodysplastic syndrome/AML (particularly erythroleukemia), chronic myeloid leukemia, non-Hodgkin lymphoma (principally, follicular lymphoma [FL]), and multiple myeloma, accompanied by immune-mediated and granulomatous disorders. The p.R164W variant is notable by its predisposition to lymphoproliferative neoplasms in 5 family members, suggesting that the disease profile may, in part, be governed by the underlying germ line lesion and the timing and nature of secondary mutations. Such interfamilial and intrafamilial variation underlines the need for heightened clinical awareness to first suspect and then detect the underlying genetic predisposition. This task is made increasingly difficult by the latency of disease onset in these pedigrees with a median age of presentation across all DDX41 pedigrees of 62 years (33-74 years), contrasting with other leukemia predisposition syndromes where the majority of affected individuals present at an earlier age (<45 years). Indeed, Lewinsohn and colleagues suggest that missense DDX41 mutations may identify a particular cohort of patients with a relatively younger age of disease onset, as noted by the occurrence of FL in 3 patients all <55 years at diagnosis.
Collectively, these findings highlight the need to integrate this gene into current diagnostic algorithms. In broader terms, perhaps 1 of the key lessons arising from these studies is the importance of collaboration to maximize research efforts within this rare patient population and to promote increased vigilance on behalf of the wider hematology community. Critically, with the advent of NGS technologies, multicenter studies have led to a rapid increase in our understanding of the genetic complexity of inherited HM, with the number of susceptibility loci more than doubling in the last 3 years. These emerging data are essential to enable comprehensive investigation and tailored long-term management of patients and their families, while continuing to offer novel insights into the molecular pathogenesis of HM.
Conflict-of-interest disclosure: The authors declare no competing financial interests.