In this issue of Blood, Elena et al1 report integration of molecular genetic data into a previously published2 chronic myelomonocytic leukemia (CMML)–specific prognostic scoring system (CPSS); the resulting clinical/molecular CPSS (CPSS-Mol) is a new 4-tier integrated clinical-pathological-genetic risk-stratification tool for CMML.
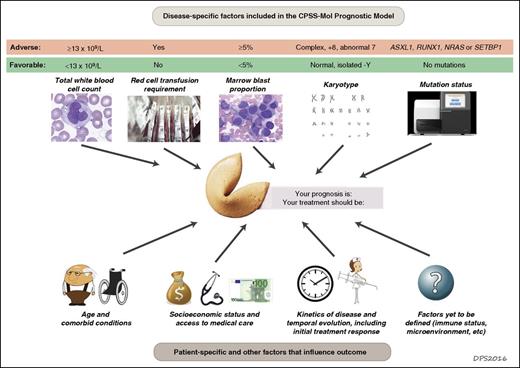
Prognosis and risk assessment in CMML. Prognostication for patients with CMML requires integration of diverse data. Adverse disease-specific factors incorporated into the new CPSS-Mol risk-stratification tool include a high white cell count (a marker of the proliferative capacity of the dominant hematopoietic clone), red cell transfusion dependency (a marker of the degree of marrow failure), increased marrow blast proportion, high risk karyotype, and somatic mutations in ASXL1, NRAS, SETBP1, or RUNX1. Other factors will also influence patient outcome, including older age, more severe comorbid conditions with poorer performance status, low socioeconomic status, poor access to high-quality health care, rapid disease progression, and failure to respond to therapy. It seems likely that additional factors that are currently more difficult to measure also influence clinical outcomes, such as the ability of the patient’s immune system to restrain clonal outgrowth, epigenetic patterns, the repertoire of aberrant splicing isoforms, or interactions of the clone with the marrow microenvironment.
Prognosis and risk assessment in CMML. Prognostication for patients with CMML requires integration of diverse data. Adverse disease-specific factors incorporated into the new CPSS-Mol risk-stratification tool include a high white cell count (a marker of the proliferative capacity of the dominant hematopoietic clone), red cell transfusion dependency (a marker of the degree of marrow failure), increased marrow blast proportion, high risk karyotype, and somatic mutations in ASXL1, NRAS, SETBP1, or RUNX1. Other factors will also influence patient outcome, including older age, more severe comorbid conditions with poorer performance status, low socioeconomic status, poor access to high-quality health care, rapid disease progression, and failure to respond to therapy. It seems likely that additional factors that are currently more difficult to measure also influence clinical outcomes, such as the ability of the patient’s immune system to restrain clonal outgrowth, epigenetic patterns, the repertoire of aberrant splicing isoforms, or interactions of the clone with the marrow microenvironment.
CMML has long been challenging to classify and difficult to manage.3 This heterogeneous group of diseases is currently defined by the coexistence of myeloproliferative neoplasm (MPN)–associated features (eg, leukocytosis or organomegaly), myelodysplastic syndromes (MDS)–associated features (eg, marrow failure and cell dysmorphology), and a blood monocyte count of ≥1 × 109/L. CMML was considered 1 of 5 forms of MDS by the 1982 French-American-British (FAB) MDS classification, in part because proliferative features are often so minimal that other than modest blood monocytosis crossing the arbitrary disease-defining threshold, CMML is indistinguishable from typical MDS. But in other respects, CMML fit with the other “FAB 4” syndromes about as well as Yoko Ono fit with The Beatles, so in 2001 the World Health Organization broke up the FAB group and relegated CMML to a separate cluster of MDS/MPN overlap neoplasms, of which CMML is by far the most commonly encountered entity.
Limited biological understanding of CMML and a paucity of effective treatments have historically hampered progress, resulting in poor outcomes for most patients. Only a single CMML-specific randomized trial has ever been completed: 20 years ago, a French study demonstrated the superiority of hydroxyurea to oral etoposide.4
More recently, however, a clearer picture of the unique biology of CMML has begun to emerge, including the overrepresentation of certain leukemic driver mutations (eg, SRSF2, ASXL1, CBL, SETBP1) in CMML compared with other myeloid neoplasms.5 Recognition of the importance of granulocyte-macrophage colony-stimulating factor (GM-CSF) signaling via the JAK-STAT pathway in CMML pathobiology6 prompted a successful MDS Clinical Research Consortium pilot trial of the JAK1/2 inhibitor ruxolitinib in CMML,7 and MDS/MPN overlap-specific consensus response criteria were recently proposed.8 A second randomized CMML-specific French trial is ongoing, this time of decitabine vs hydroxyurea (NCT02214407). And investigators are also proposing useful new prognostic models.
As is the case for MDS without proliferative features, disease heterogeneity in CMML means that risk-stratification tools are helpful to counsel patients and make informed decisions about the appropriate intensity of therapy. Since the 1980s, various groups have developed CMML prognostic tools based on clinical and pathological criteria, including the CPSS. Several risk-stratification tools integrating molecular findings have been developed more recently, including the Mayo Molecular Model and the Groupe Francophone des Myélodysplasies (GFM) Model.9,10 A consistent theme across the newer models has been that anemia, leukocytosis, and ASXL1 mutations are independent negative prognostic factors.
The newest tool, CPSS-Mol, was derived from multivariable modeling of a learning cohort of 214 Italian, German, and Spanish patients, 93% of whom had a mutation in at least 1 of 38 leukemia-associated genes. The model was then validated in an independent cohort of 260 patients. CPSS-Mol includes 5 features: red cell transfusion dependence, white blood cell count (previously used to separate “proliferative” from “dysplastic” CMML subtypes), marrow blast proportion, karyotype, and mutation status for 4 genes (see figure). When the team that derived CPSS-Mol compared it to the other models that incorporate molecular genetic data, it performed better than either the Mayo or GFM models; other investigators will need to validate this result in independent data sets. The huge difference in median survival between CPSS-Mol risk groups, from 18 months in the highest risk cohort to >144 months in the lowest, underscores the remarkable heterogeneity of CMML.
Prognostic systems are most helpful if they allow clinicians to choose 1 treatment path over another. Currently, however, the choice of paths for patients with CMML is limited, as the menu of effective drugs is short. Clinicians treating patients with CMML most commonly use either a hypomethylating agent (decitabine or azacitidine), a myelosuppressive drug such as hydroxyurea, cytarabine, or cladribine, or a hematopoietic growth factor. Outcomes with reduced-intensity conditioning allogeneic stem cell transplant tend to be poor in MDS/MPN overlap syndromes such as CMML because the proliferative clone regrows quickly, before a graft-versus-leukemia effect can develop. A few CMML-specific trials of new agents are ongoing, including studies of the farnesyltransferase inhibitor tipifarnib, the hypomethylating agent guadecitabine, and the anti–GM-CSF drug KB003. As better CMML risk-stratification tools such as CPSS-Mol are developed, we can hope that they will be accompanied by novel therapies that can improve outcomes for all of the risk groups defined by these tools.
Conflict-of-interest disclosure: The author declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal