Key Points
EBV LMP1 dysregulates EphA4 expression via the ERK-Sp1 pathway.
Downregulation of EphA4 is demonstrated in EBV+ DLBCL, which is significantly correlated with poor DLBCL survival.
Abstract
Epstein-Barr virus (EBV), an oncogenic human virus, is associated with several lymphoproliferative disorders, including Burkitt lymphoma, Hodgkin disease, diffuse large B-cell lymphoma (DLBCL), and posttransplant lymphoproliferative disorder (PTLD). In vitro, EBV transforms primary B cells into lymphoblastoid cell lines (LCLs). Recently, several studies have shown that receptor tyrosine kinases (RTKs) play important roles in EBV-associated neoplasia. However, details of the involvement of RTKs in EBV-regulated B-cell neoplasia and malignancies remain largely unclear. Here, we found that erythropoietin-producing hepatocellular receptor A4 (EphA4), which belongs to the largest RTK Eph family, was downregulated in primary B cells post-EBV infection at the transcriptional and translational levels. Overexpression and knockdown experiments confirmed that EBV-encoded latent membrane protein 1 (LMP1) was responsible for this EphA4 suppression. Mechanistically, LMP1 triggered the extracellular signal-regulated kinase (ERK) pathway and promoted Sp1 to suppress EphA4 promoter activity. Functionally, overexpression of EphA4 prevented LCLs from proliferation. Pathologically, the expression of EphA4 was detected in EBV− tonsils but not in EBV+ PTLD. In addition, an inverse correlation of EphA4 expression and EBV presence was verified by immunochemical staining of EBV+ and EBV− DLBCL, suggesting EBV infection was associated with reduced EphA4 expression. Analysis of a public data set showed that lower EphA4 expression was correlated with a poor survival rate of DLBCL patients. Our findings provide a novel mechanism by which EphA4 can be regulated by an oncogenic LMP1 protein and explore its possible function in B cells. The results provide new insights into the role of EphA4 in EBV+ PTLD and DLBCL.
Introduction
Epstein-Barr virus (EBV) is oncogenic and its infection is associated with multiple human diseases, especially lymphoproliferative disorders, such as infectious mononucleosis, Burkitt lymphoma (BL), Hodgkin disease, natural killer/T-cell lymphoma, posttransplant lymphoproliferative disorder (PTLD), and EBV+ diffuse large B-cell lymphoma (DLBCL).1,2 In vitro, EBV can transform human primary B cells into lymphoblastoid cell lines (LCLs), which support its association with human malignancies. Like other human herpes viruses, EBV may be found in 2 states: latency and lytic replication. It is well documented that viral latent products contribute largely to EBV oncogenic activities. The main EBV latent products include 6 EBV nuclear antigens (EBNA1, 2, LP, 3A, 3B, 3C), 3 latent membrane proteins (LMP1, 2A, 2B), 2 EBV-encoded small RNAs (EBER1, 2), and microRNAs (Bam H1 rightwards transcripts).1 According to previous studies, EBV uses various strategies to manipulate cell gene expression so that it can persist in infected B cells. These include altering the expression of protein tyrosine kinases (PTKs), cytokines, adhesion molecules, and antiapoptotic genes.1,3-8 Among them, we are particularly interested in PTKs because they are important at many aspects of cell proliferation, differentiation, apoptosis, migration, and tissue development. In total, 90 PTKs have been defined following sequencing of the human genome.9 They can be divided into 2 major categories: 58 receptor tyrosine kinases (RTKs) and 32 non-RTKs.9 In the case of EBV-associated malignancies, several EBV gene products are involved in dysregulation of RTKs. EBV-encoded LMP1 elevates amounts of Recepteur d’Origine Nantais via NF-κB to promote B-cell proliferation and induce the migration and invasion of epithelial cells.3,4 In addition, epidermal growth factor receptor is induced and activated by LMP1 through protein kinase Cδ and NF-κB.10-12 Also, c-Met is upregulated by LMP1 via Ets1 in epithelial cells.13 Activation of discoidin domain receptor 1 by LMP1 contributes to the survival of Hodgkin lymphoma.14 Recently, LMP1 has been reported to increase the expression of fibroblast growth factor receptor 1, enhancing aerobic glycolysis and cell invasiveness.15 Furthermore, LMP1 activates the insulin-like growth factor 1 receptor via upregulation of its ligand, IGF1, to promote cell proliferation and colony formation.16 In addition, Zta, a lytic EBV transactivator, can upregulate the expression of Trk-related tyrosine kinase, which may enhance the metastasis of nasopharyngeal carcinoma.5,6 Human epidermal growth factor receptor 2 (HER2) and HER3 are dysregulated by EBV-encoded BARF0, which not only increases the level of HER2/3 but also enhances cell-anchorage independence.17 In the cases mentioned above, many EBV products are responsible for increasing the expression or enhancing the kinase activity of RTKs to deliver signaling that favors cell proliferation, migration, or invasion. Herein, we wondered whether there are other mechanisms involved in EBV immortalization of B cells through other RTKs. In our previous study, a kinase display was used to investigate the differential expression of PTKs between primary B cells and LCL. Recepteur d’Origine Nantais and Trk-related tyrosine kinase were found to be upregulated.3-6 In this manuscript, we reveal another RTK, namely erythropoietin-producing hepatocellular receptor A4 (EphA4), which is involved in EBV lymphoproliferation.
Of note, Eph receptor is the largest family of RTKs in humans, being composed of 9 EphAs and 5 EphBs. Usually, EphA receptors bind to glycosylphosphatidylinositol-linked Eph receptor-interacting protein (ephrin)-A ligands, and Eph B receptors bind to transmembrane ephrin-B ligands.18,19 The interactions between Eph receptors and their ligands trigger both forward Eph kinase activity in Eph receptor-expressing cells and reverse Src family kinase activity in ephrin-expressing cells.19 These bidirectional signaling pathways make the Eph family distinct from other RTKs. Conventionally, their functions are recognized in developmental processes and tissue homeostasis.20 Recently, dysregulation of Eph receptors and ephrins has been reported in many human cancers.18,19,21,22 Of interest, various reports indicated that Eph can function in oncogenic promotion or tumor suppression in several human cancers at different stages or in different cancers.18,19,21,22 Notably, the involvement of Eph in viral infection has been explored in some human oncogenic viruses. EphA2 is the entry receptor for Kaposi sarcoma-associated herpesvirus23 and also acts with epidermal growth factor receptor as a coreceptor for hepatitis C virus entry.24 Collectively, Ephs not only play roles in physiological activities but also are involved in pathogenesis.25 According to our kinase display data, EphA4 is clearly downregulated in LCL, compared with uninfected B cells. Little is known about the role of EphA4 in B lymphocytes, so the expression and regulation of EphA4 in LCL and EBV-associated diseases will be explored in this article.
Materials and methods
B-cell purification and EBV infection
Peripheral blood mononuclear cells were kindly provided by anonymous donors at Taipei Blood Center of Taiwan Blood Service Foundation, and CD19+ B cells were purified as described previously.7 The methods of EBV virion (B95.8 strain) production and EBV infection of primary B cells have been described previously.7 Experiments using human samples were approved by institutional review boards of National Taiwan University Hospital, Taipei, Taiwan.
Cell culture
Human peripheral blood CD19+ B cells were infected by B95.8 EBV to establish LCLs in vitro. BJAB cells are EBV− BL cells. All B-cell lines were cultured in complete RPMI medium containing 10% fetal calf serum, 1 mM glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin. A nasopharyngeal carcinoma cell line, TW01, was maintained in complete Dulbecco modified Eagle medium containing 10% fetal calf serum, 1 mM glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin.
Kinase display assay
The tyrosine kinase display assays were performed as described previously.5 Briefly, total RNAs were extracted and complementary DNAs (cDNAs) of PTKs were generated and amplified by using [γ-33P]-labeled degenerate primers designed for the conserved kinase domain. The resulting 170-bp polymerase chain reaction (PCR) products were gel-purified and digested separately using 16 restriction enzymes. The digested PCR products were resolved in a denaturing polyacrylamide gel. The patterns of restriction enzyme digestion were identified for each specific PTK in the data bank.
Plasmids
The EBNA1-, EBNA2-, Zta-, and LMP1-expressing plasmids were constructed as described previously.7 The luciferase reporter plasmid, driven from the EPHA4 promoter (nucleotides −1000 to +42), was inserted into the pGL3-basic vector (Promega). The LMP1-expressing lentivirus plasmid, pSIN-LMP1, and its mutant plasmids, including LMP1-deleted C-terminal activation region 1 (CTAR1; ΔCTAR1) with the deletion of LMP1 amino acids 194 to 232, deleted CTAR2 (ΔCTAR2) with deletion of LMP1 amino acids 351 to 386 and LMP1-deleted CTAR1/2 (ΔCTAR1/2) plasmids were reported previously.3 The shLMP1, shSp1 lentivirus plasmids and its vector control shLuc were constructed in the pLKO.1 plasmids as described previously.3,26 The pLKO.1-shERK1/2 lentivirus plasmids were purchased from National RNAi Core Facility (Academia Sinica, Taipei, Taiwan) with sequences targeting human extracellular signal-regulated kinase 1#1 (ERK1#1) (5′-CTATACCAAGTCCATCGACAT-3′), ERK1#2 (5′-TCCCTGTCAAAGCTGTCACTT-3′), and ERK2 (5′-TATCCATTCAGCTAACGTTCT-3′). The EphA4 (pCDNA3.1-EphA4) full-length (wild-type [WT]) and mutant plasmids are kindly provided by Dr Tang-Long Shen (Department of Plant Pathology and Microbiology, National Taiwan University, Taipei, Taiwan). The Flag-tagged EphA4 WT and mutant lentivirus plasmids were constructed into pWPI lentiviral plasmid, which is a gift from Dr Min-Liang Kuo (Institute of Toxicology, National Taiwan University, Taipei, Taiwan).
RNA extraction, RT-Q-PCR
Total RNAs were extracted by using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Reverse transcription and quantitative PCR (RT-Q-PCR) has been described previously.26 EphA4 messenger RNA (mRNA) was determined using forward primer 5′-GATAGCAAGCCCTCTGGAAG-3′, reverse primer 5′-CCAATCAGTTCGTAGCCAGTT-3′, and Roche no. 20 universal probe. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA were determined by using forward primer 5′-TCCACTGGCGTCTTCACC-3′, reverse primer 5′-GGCAGAGATGATGACCCTTTT-3′, and Roche no. 45 universal probe. MAGOH mRNA were determined by using forward primer 5′-AAAGAGGATGATGCATTGTGGt-3′, reverse primer 5′-TCTTCTCCAATGACGATTTCA T-3′, and Roche no. 51 universal probe.
Western blotting and antibodies
Cells were lysed using radioimmunoprecipitation assay buffer and western blotting was performed as per previous study.7 Antibodies (Abs) were used as follows: EphA4 (Santa Cruz Biotechnology and ECM Biosciences), β-actin (Sigma-Aldrich), phospho-Akt S473 (Cell Signaling), Akt (Santa Cruz Biotechnology), phospho-IκB-α-S32/36 (Cell Signaling), p65 (Santa Cruz Biotechnology), phospho-JNK T183/Y185 (Cell Signaling), JNK (Millipore), phospho-ERK T202/Y204 (Cell Signaling), ERK (Santa Cruz Biotechnology), Sp1 (Santa Cruz Biotechnology). Abs against EBV viral products including EBNA1, EBNA2, LMP1, Zta were as reported previously.7,27
Infection of lentivirus
The method of production and infection with lentivirus was reported previously.7 For EphA4 WT and mutant-expressing lentivirus infection, 5 × 105 LCLs were infected with lentivirus at a multiplicity of infection (MOI) of 4. For LMP1, Sp1, and ERK1/2 knockdown, LCLs were seeded at a density of 1 × 106 cells/mL and infected with shLuc control, shLMP1, shSp1, or shERK1/2 lentivirus at MOI of 1. BJAB cells were seeded in 1 × 106 cells/mL and infected with pSIN-LMP1 and its deletion mutant expressing lentivirus at a range of 0.5 to 4 MOI.
Treatment with inhibitors
LCLs were seeded at 1 × 106 cells/mL and treated with 20 μM phosphatidylinositol 3-kinase (PI3K) inhibitor LY294002, 20 μM JNK inhibitor SP600125, 20 μM MEK inhibitor PD98059, 2.5 μM NF-κB inhibitor BAY 11-7082 for 48 hours, respectively. TW01 cells were treated with 500 nM Sp1 inhibitor mithramycin for 24 hours. The inhibitors mentioned above were purchased from Merck Millipore. Dimethylsulfoxide served as a solvent control.
Reporter assay
TW01 cells were seeded at a density of 1 × 105 cells per well in a 12-well plate and then cotransfected with 0.5 μg of EPHA4 promoter luciferase reporter plasmids, 0.25 μg of pSG5-LMP1, and 0.05 μg of green fluorescent protein (GFP)-expressing plasmids (pEGFP-C1; Promega) using T-Pro Non-liposome transfection Reagent II (T-Pro NTR II; T-Pro Biotechnology) according to the manufacturer’s instructions. Cells were harvested and the luciferase activities and GFP fluorescent intensity were detected using the Bright-Glo Luciferase Assay System kit (Promega) 2 days posttransfection. The relative fold induction of luciferase activity from each transfectant was first standardized with enhanced GFP, followed by normalization to the control vector pGL3.
Proliferation assay
Lentivirus-infected LCLs were seeded at 1 × 104 cells per well in 96-well plates for 5 days. Before the indicated time point, cells were treated with AlamarBlue (Thermo Fisher Scientific) for 4 hours and the absorbance measured according to the manufacturer’s instructions.
Chromatin immunoprecipitation assay
BJAB vector control and LMP1-expressing cells were harvested. DNA-protein complexes were immunoprecipitated using anti-Sp1 antibody as described previously.28 The DNA was extracted and analyzed by PCR with the EPHA4 promoter spanning the Sp1-binding sites. The amplification of the GAPDH promoter region was as described previously.7
IHC and in situ hybridization of EBER assays
Tonsil, PTLD, and DLBCL biopsies were obtained from the National Taiwan University Hospital. Immunohistochemistry (IHC) assays of EphA4 and LMP1 were performed using the Starr Trek Universal Detection system (Biocare) according to the manufacturer’s protocol, and in situ hybridization of EBER assays was described in our previous study.3
Kaplan-Meier plots
The microarray data sets of DLBCL were downloaded from the National Center for Biotechnology Information (NCBI) and Gene Expression Omnibus (GEO) database (accession number GSE4475). Overall survival was analyzed by generating Kaplan-Meier plots. Patients were divided into high and low EphA4 groups, according to the median expression of EphA4.
Statistical analysis
Statistical analysis was conducted using GraphPad Prism software program. Quantitative data were reported as mean ± standard error of the mean and compared using the unpaired Student t test. Clinical correlation was determined by Fisher exact test. For Kaplan-Meier survival analysis, a log-rank test was used to compare the difference between 2 groups. Data were considered statistically significant at P < .05.
Results
EphA4 is downregulated post-EBV infection
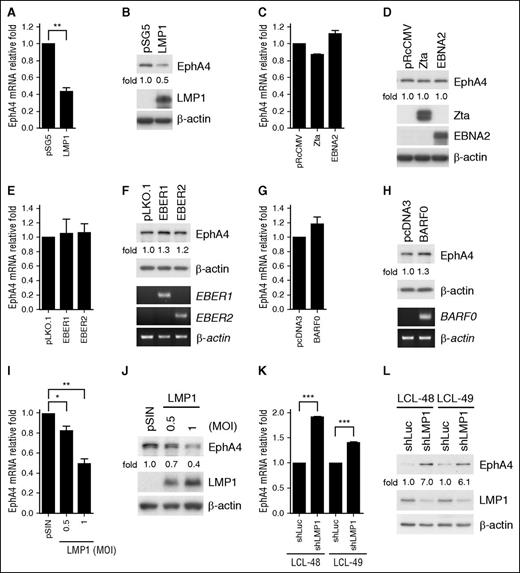
Because RTKs play a critical role in EBV-infected cells, the RTK expression profile was analyzed by kinase display assay using degenerate primers.5 According to the BstNI, HhaI, and MnlI restriction enzyme digestion profile, downregulation of EphA4 was found in 3 LCLs compared with control primary B cells (Figure 1A). To confirm this result, mRNA and protein expression levels of EphA4 were detected in cells infected with EBV for 3, 7, 14, 21, or 28 days (defined as LCL), compared with uninfected primary B cells. Clearly, mRNA and protein expression levels of EphA4 were reduced to ∼50% to 10% post-EBV infection, compared with primary B cells (Figure 1B-C). Expression of EBNA1 and LMP1 indicated that the primary B cell was successfully transformed by EBV (Figure 1C). In addition to EBV infection, B cells can be activated by various treatments including anti-CD40 antibody plus interleukin-4 (IL-4), lipopolysaccharide (LPS), and poly I:C to mimic T-cell–dependent or –independent activation.29 To clarify whether B-cell activation can decrease EphA4 expression, EphA4 transcripts were measured after 3 days of incubation. As shown in Figure 1D, EphA4 was downregulated not only by EBV infection but also by anti-CD40 antibody plus IL-4 treatment. This fit previous findings that EBV LMP1 functionally mimics the constitutively active CD40 receptor associated with tumor necrosis factor receptor-associated factors.30,31 Meanwhile, treatments of LPS or poly I:C which triggered the Toll-like receptor 4 or 3 signal pathway did not alter EphA4 expression.
EphA4 expression is decreased post-EBV infection. (A) Total RNAs were harvested from uninfected primary B cells, from peripheral blood, and EBV-immortalized LCLs from 3 different donors. cDNAs were generated using [33P]-labeled degenerate primers for PTKs. EphA4-specific cDNAs were digested by 3 restriction enzymes, BstNI, HhaI, and MnlI. Relative fold EphA4 expression was compared with primary B cells. (B-C) Peripheral CD19+ B cells were seeded at 1 × 106 cells/mL and infected with EBV strain B95.8. Total RNAs and protein were harvested from primary B cells at the days indicated post-EBV infection. (B) Expression levels of EphA4 mRNA were measured by RT-Q-PCR. EphA4 mRNA-relative folds were normalized to internal control MAGOH and standardized with uninfected primary B cells. (C) EphA4, EBNA1, LMP1, and β-actin proteins were measured by western blotting. EphA4 protein-relative folds were normalized to internal control β-actin and compared with uninfected primary B cells. (D) Total RNAs were extracted from primary B cells, EBV infection, or B-cell stimulations including anti-CD40 antibody plus IL-4, LPS, or poly I:C for 3 days. EphA4 transcripts were measured by RT-Q-PCR. EphA4 mRNA-relative folds were normalized to internal control MAGOH and standardized with uninfected primary B cells. (E) Total RNAs were extracted from primary B cells and 8 LCL lines. Expression levels of EphA4 mRNA were measured by RT-Q-PCR. EphA4 mRNA-relative folds were normalized to internal control MAGOH and standardized with uninfected primary B cells. (F-G) Total RNAs and protein were harvested from paired uninfected B cells and LCLs generated from the peripheral blood mononuclear cells of 2 healthy donors. (F) Expression levels of EphA4 mRNA were measured by RT-Q-PCR. EphA4 mRNA-relative folds were normalized to internal control MAGOH and standardized with uninfected primary B cells. (G) EphA4, EBNA1, LMP1, and β-actin proteins were measured by western blotting. EphA4 protein-relative folds were normalized to β-actin and compared with uninfected primary B cells.
EphA4 expression is decreased post-EBV infection. (A) Total RNAs were harvested from uninfected primary B cells, from peripheral blood, and EBV-immortalized LCLs from 3 different donors. cDNAs were generated using [33P]-labeled degenerate primers for PTKs. EphA4-specific cDNAs were digested by 3 restriction enzymes, BstNI, HhaI, and MnlI. Relative fold EphA4 expression was compared with primary B cells. (B-C) Peripheral CD19+ B cells were seeded at 1 × 106 cells/mL and infected with EBV strain B95.8. Total RNAs and protein were harvested from primary B cells at the days indicated post-EBV infection. (B) Expression levels of EphA4 mRNA were measured by RT-Q-PCR. EphA4 mRNA-relative folds were normalized to internal control MAGOH and standardized with uninfected primary B cells. (C) EphA4, EBNA1, LMP1, and β-actin proteins were measured by western blotting. EphA4 protein-relative folds were normalized to internal control β-actin and compared with uninfected primary B cells. (D) Total RNAs were extracted from primary B cells, EBV infection, or B-cell stimulations including anti-CD40 antibody plus IL-4, LPS, or poly I:C for 3 days. EphA4 transcripts were measured by RT-Q-PCR. EphA4 mRNA-relative folds were normalized to internal control MAGOH and standardized with uninfected primary B cells. (E) Total RNAs were extracted from primary B cells and 8 LCL lines. Expression levels of EphA4 mRNA were measured by RT-Q-PCR. EphA4 mRNA-relative folds were normalized to internal control MAGOH and standardized with uninfected primary B cells. (F-G) Total RNAs and protein were harvested from paired uninfected B cells and LCLs generated from the peripheral blood mononuclear cells of 2 healthy donors. (F) Expression levels of EphA4 mRNA were measured by RT-Q-PCR. EphA4 mRNA-relative folds were normalized to internal control MAGOH and standardized with uninfected primary B cells. (G) EphA4, EBNA1, LMP1, and β-actin proteins were measured by western blotting. EphA4 protein-relative folds were normalized to β-actin and compared with uninfected primary B cells.
To determine whether this effect is common in B cells infected by EBV, expression of EphA4 mRNA was detected by RT-Q-PCR in another 8 LCL cell lines established in our laboratory.7 Compared with primary B cells, EphA4 transcripts were also detected at 10% to 50% normal levels in LCLs (Figure 1E). To verify these observations, total RNA and protein were collected from 2 paired primary B cells and their corresponding LCLs. Consistent with Figure 1B-C, EphA4 mRNA and protein expression were reduced in LCLs, suggesting that EBV infection influenced the expression of EphA4 at the transcriptional and translational levels (Figure 1F-G).
LMP1 is responsible for EphA4 suppression
Many Epstein-Barr viral products are able to regulate gene expression. To determine which Epstein-Barr viral gene was involved in EphA4 suppression, selected viral genes were expressed ectopically in the EBV− nasopharyngeal carcinoma cell TW01 through transfection. Based on the results of Figure 2A-H, we demonstrated that LMP1 was the candidate protein that inhibits EphA4 expression. Other EBV gene products, including Zta, EBNA2, EBER1, EBER2, and BARF0, had no effect on the EphA4 expression level (Figure 2C-H). This LMP1-mediated EphA4 downregulation was also observed in a dose-dependent manner in EBV− BL cell BJAB transduced with LMP1 lentivirus, at mRNA and protein levels (Figure 2I-J). Of note, upregulation of EphA4 protein and mRNA expression was seen in LCLs using an shLMP1 knockdown approach (Figure 2K-L). Taken together, all of the evidence indicated that LMP1 was the viral protein responsible for EphA4 downregulation.
EphA4 is downregulated by LMP1. (A-H) EBV− TW01 cells were transfected with plasmids harboring the EBV viral genes LMP1, Zta, EBNA2, EBER1, EBER2, and BARF0. Total RNAs and protein lysates were obtained from each transfectant at day 3 posttransfection. (A, C, E, G) EphA4 transcripts were detected by RT-Q-PCR. EphA4 mRNA-relative folds were normalized to internal control GAPDH and then standardized with vector controls. (Top panels of B, D, F, H) Total proteins were harvested from the vector control and each transfectant. EphA4 protein-relative folds were normalized to internal control β-actin and standardized with vector controls. Expression levels of LMP1, Zta, EBNA2, and β-actin protein were estimated by western blotting. (Bottom panels of F and H) The EBER1, EBER2, BARF0, and β-actin transcripts were analyzed by RT-PCR. (I-J) EBV− BJAB cells were infected with LMP1-expressing lentivirus at MOI 0.5 or 1. The expression levels of EphA4 mRNA were determined by RT-Q-PCR. EphA4 mRNA-relative folds were normalized to internal control GAPDH and then standardized with the pSIN vector control. (J) EphA4, LMP1, and β-actin were detected by western blotting. EphA4 protein-relative folds were normalized to internal control β-actin and then standardized with pSIN vector control. (K-L) Knockdown of LMP1 in LCLs was performed by lentiviral transduction at the MOI of 1 for 5 days and infected cells were selected with 2 µg/mL puromycin for 2 days. (K) Expression levels of EphA4 mRNA were measured by RT-Q-PCR. EphA4 mRNA-relative folds were normalized to internal control GAPDH and then standardized with vector control shLuc. (L) EphA4, LMP1, and β-actin were determined by western blotting. EphA4 protein-relative folds were normalized to β-actin and then standardized with vector control shLuc (*P < .05; **P < .01; ***P < .001, Student t test).
EphA4 is downregulated by LMP1. (A-H) EBV− TW01 cells were transfected with plasmids harboring the EBV viral genes LMP1, Zta, EBNA2, EBER1, EBER2, and BARF0. Total RNAs and protein lysates were obtained from each transfectant at day 3 posttransfection. (A, C, E, G) EphA4 transcripts were detected by RT-Q-PCR. EphA4 mRNA-relative folds were normalized to internal control GAPDH and then standardized with vector controls. (Top panels of B, D, F, H) Total proteins were harvested from the vector control and each transfectant. EphA4 protein-relative folds were normalized to internal control β-actin and standardized with vector controls. Expression levels of LMP1, Zta, EBNA2, and β-actin protein were estimated by western blotting. (Bottom panels of F and H) The EBER1, EBER2, BARF0, and β-actin transcripts were analyzed by RT-PCR. (I-J) EBV− BJAB cells were infected with LMP1-expressing lentivirus at MOI 0.5 or 1. The expression levels of EphA4 mRNA were determined by RT-Q-PCR. EphA4 mRNA-relative folds were normalized to internal control GAPDH and then standardized with the pSIN vector control. (J) EphA4, LMP1, and β-actin were detected by western blotting. EphA4 protein-relative folds were normalized to internal control β-actin and then standardized with pSIN vector control. (K-L) Knockdown of LMP1 in LCLs was performed by lentiviral transduction at the MOI of 1 for 5 days and infected cells were selected with 2 µg/mL puromycin for 2 days. (K) Expression levels of EphA4 mRNA were measured by RT-Q-PCR. EphA4 mRNA-relative folds were normalized to internal control GAPDH and then standardized with vector control shLuc. (L) EphA4, LMP1, and β-actin were determined by western blotting. EphA4 protein-relative folds were normalized to β-actin and then standardized with vector control shLuc (*P < .05; **P < .01; ***P < .001, Student t test).
LMP1 suppresses EphA4 through ERK pathways
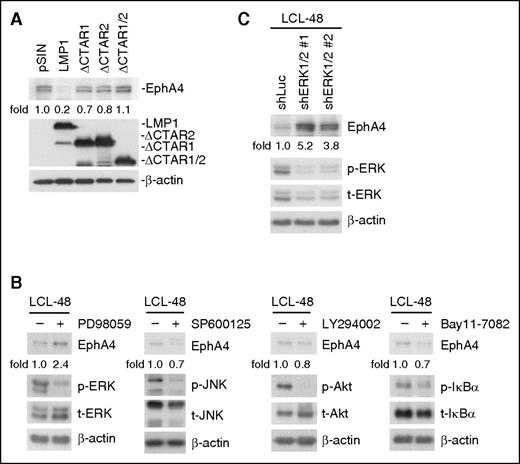
Structurally, LMP1 resembles a CD40 receptor but its activity is ligand-independent. LMP1 delivers signaling through its long CTARs.30,31 To determine which CTAR domains were involved in EphA4 downregulation, lentiviruses containing LMP1 full-length (WT), CTAR1-deleted mutant (ΔCTAR1), CTAR2-deleted mutant (ΔCTAR2), and both CTAR1- and CTAR2-deleted mutant (ΔCTAR1/2) were transduced into BJAB cells. It seems that both CTAR1 and 2 of LMP1 are involved in this repression (Figure 3A). Usually, these CTARs associate with tumor necrosis factor receptor-associated factors and activate downstream PI3K/Akt, NF-κB, and MAPK, including the JNK, ERK, and p38 pathways.32 To dissect which signaling pathway was necessary for LMP1-mediated EphA4 downregulation, LCLs were treated with MEK inhibitor PD98059, JNK inhibitor SP600125, PI3K inhibitor LY294002, or NF-κB inhibitor Bay11-7082. Expression of EphA4 was increased in LCLs treated with PD98059 but not other inhibitors (Figure 3B). Furthermore, LMP1-triggered EphA4 downregulation through the ERK pathway was confirmed by silencing both ERK1/2 by lentiviral transduction of LCLs (Figure 3C).
The ERK pathway is critical for LMP1 mediation of EphA4 downregulation. (A) BJAB cells were infected with pSIN-, LMP1-, ΔCTAR1-, ΔCTAR2-, or ΔCTAR1/2-expressing lentiviruses for 5 days. EphA4, LMP1, and β-actin were detected by western blotting. EphA4 protein-relative folds were normalized to β-actin and standardized with pSIN vector control. (B) LCLs were treated with 20 μM PD98059, SP600125, LY294002, or 2.5 μM Bay11-7082 for 48 hours. EphA4, phosphorylated (p-) and total (t-) proteins of ERK, JNK, Akt, and IkBα were determined by western blotting. β-actin served as an internal control. EphA4 protein-relative folds were normalized to β-actin and standardized with dimethylsulfoxide solvent control. (C) Knockdown of ERK1/2 in LCLs achieved using a lentivirus expressing shERK1 plus shERK2 for 3 days. Infected cells were selected with 2 µg/mL puromycin for 2 days. EphA4, phospho-ERK, total ERK proteins were then determined by western blotting. EphA4 protein-relative folds were normalized to β-actin and standardized with vector control shLuc.
The ERK pathway is critical for LMP1 mediation of EphA4 downregulation. (A) BJAB cells were infected with pSIN-, LMP1-, ΔCTAR1-, ΔCTAR2-, or ΔCTAR1/2-expressing lentiviruses for 5 days. EphA4, LMP1, and β-actin were detected by western blotting. EphA4 protein-relative folds were normalized to β-actin and standardized with pSIN vector control. (B) LCLs were treated with 20 μM PD98059, SP600125, LY294002, or 2.5 μM Bay11-7082 for 48 hours. EphA4, phosphorylated (p-) and total (t-) proteins of ERK, JNK, Akt, and IkBα were determined by western blotting. β-actin served as an internal control. EphA4 protein-relative folds were normalized to β-actin and standardized with dimethylsulfoxide solvent control. (C) Knockdown of ERK1/2 in LCLs achieved using a lentivirus expressing shERK1 plus shERK2 for 3 days. Infected cells were selected with 2 µg/mL puromycin for 2 days. EphA4, phospho-ERK, total ERK proteins were then determined by western blotting. EphA4 protein-relative folds were normalized to β-actin and standardized with vector control shLuc.
LMP1 represses EphA4 promoter activity through Sp1
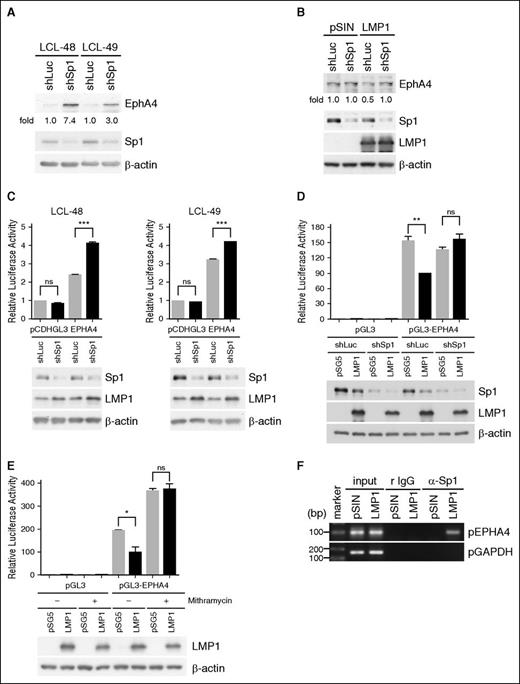
LMP1 downregulated the expression of EphA4 at the transcriptional and translational levels (Figure 2) through the ERK pathway (Figure 3B-C). ERK has been reported to phosphorylate Sp1 at T453 and T739 to regulate the promoter activity of targeted genes33 and we speculated that LMP1 may inhibit EphA4 promoter activity through Sp1. To test this hypothesis, EphA4 protein was detected in Sp1-knockdown LCLs and its amount was augmented (Figure 4A). Meanwhile, EphA4 protein amounts were restored in LMP1-overexpressing BJAB cells, when cells were simultaneously knocked down of Sp1 (Figure 4B). These data suggested that Sp1 was involved in LMP1-mediated EphA4 downregulation. The effect of Sp1 on EphA4 promoter activity was investigated further. The EPHA4 promoter (−1000 to +42) sequence was analyzed and 4 Sp1-binding sites were predicted. Clearly, EphA4 promoter activity was upregulated in Sp1-silenced LCLs and in Sp1-knockdown, LMP1-expressing TW01 cells (Figure 4C-D). Also, addition of the Sp1 inhibitor mithramycin, which interferes with Sp1 binding to the guanine-cytosine sequence, to the LMP1 transfectants increased the EphA4 promoter activity (Figure 4E). Furthermore, to determine whether LMP1 promotes Sp1 binding to the EPHA4 promoter, a chromatin immunoprecipitation assay was performed in BJAB cells expressing LMP1. We showed that Sp1 bound to the EPHA4 promoter at the region of −51 to −60 nt in BJAB cells, following LMP1 expression (Figure 4F).
Sp1 is the key suppressor of LMP1-hampered EPHA4 promoter activity. (A) LCLs were infected with an shSp1-expressing lentivirus for 5 days and the infected cells were selected with 2 µg/mL puromycin for 2 days. EphA4, Sp1, and β-actin expression levels were detected by western blotting. EphA4 protein-relative folds were normalized to β-actin and standardized with vector control shLuc. (B) BJAB cells were infected simultaneously with LMP1 and shSp1-expressing lentiviruses for 3 days and infected cells were selected with 2 µg/mL puromycin for 2 days. EphA4, Sp1, LMP1, and β-actin were determined by western blotting. EphA4 protein-relative folds were normalized to β-actin and standardized with pSIN plus shLuc controls. (C) LCLs were coinfected with shSp1 and GFP-tagged EPHA4 promoter (−1000 ∼ +42)-expressing lentiviruses for 3 days and then the infected cells were selected with 2 µg/mL puromycin for 2 days. EphA4-relative luciferase activity was first normalized to GFP, followed by standardization with the vector pCDHGL3 (***P < .001, Student t test). Sp1, LMP1, and internal control β-actin were analyzed by western blotting (bottom panel). (D) TW01 cells were infected with an shSp1-expressing lentivirus for 3 days and the infected cells were selected with 2 µg/mL puromycin for 2 days. The Sp1-knockdown TW01 cells were cotransfected with LMP1 plasmid or its vector control pSG5, combined with reporter plasmids EPHA4 promoter (−1000 ∼ +42) or vector control pGL3 and internal control pEGFPC1 plasmids for 2 days. EphA4-relative luciferase activity was first normalized to GFP, followed by standardization with the control vector pGL3 (**P < .01, Student t test). Expression of Sp1, LMP1, and β-actin proteins was analyzed by western blotting (bottom panel). (E) TW01 cells were cotransfected with LMP1 or pSG5 plasmids, reporter plasmids of pGL3 vector control or EPHA4 promoter (−1000 ∼ +42) and internal control pEGFPC1 plasmids. Twenty-four hours posttransfection, 500 nM mithramycin was added to the cells for another 24 hours. EphA4-relative luciferase activity was determined as described previously (*P < .05, Student t test). Expression levels of LMP1 and β-actin were measured by western blotting (bottom panel). (F) BJAB cells were transduced with pSIN- or LMP1-expressing lentiviruses for 5 days and a chromatin immunoprecipitation assay was performed as described previously. DNA-protein complexes were immunoprecipitated using anti-Sp1 Ab or isotype control rabbit immunoglobulin G (IgG). EPHA4 promoter and control GAPDH promoter DNA were detected in the immunoprecipitates by PCR. Total DNA was harvested from BJAB cells and used as the input control.
Sp1 is the key suppressor of LMP1-hampered EPHA4 promoter activity. (A) LCLs were infected with an shSp1-expressing lentivirus for 5 days and the infected cells were selected with 2 µg/mL puromycin for 2 days. EphA4, Sp1, and β-actin expression levels were detected by western blotting. EphA4 protein-relative folds were normalized to β-actin and standardized with vector control shLuc. (B) BJAB cells were infected simultaneously with LMP1 and shSp1-expressing lentiviruses for 3 days and infected cells were selected with 2 µg/mL puromycin for 2 days. EphA4, Sp1, LMP1, and β-actin were determined by western blotting. EphA4 protein-relative folds were normalized to β-actin and standardized with pSIN plus shLuc controls. (C) LCLs were coinfected with shSp1 and GFP-tagged EPHA4 promoter (−1000 ∼ +42)-expressing lentiviruses for 3 days and then the infected cells were selected with 2 µg/mL puromycin for 2 days. EphA4-relative luciferase activity was first normalized to GFP, followed by standardization with the vector pCDHGL3 (***P < .001, Student t test). Sp1, LMP1, and internal control β-actin were analyzed by western blotting (bottom panel). (D) TW01 cells were infected with an shSp1-expressing lentivirus for 3 days and the infected cells were selected with 2 µg/mL puromycin for 2 days. The Sp1-knockdown TW01 cells were cotransfected with LMP1 plasmid or its vector control pSG5, combined with reporter plasmids EPHA4 promoter (−1000 ∼ +42) or vector control pGL3 and internal control pEGFPC1 plasmids for 2 days. EphA4-relative luciferase activity was first normalized to GFP, followed by standardization with the control vector pGL3 (**P < .01, Student t test). Expression of Sp1, LMP1, and β-actin proteins was analyzed by western blotting (bottom panel). (E) TW01 cells were cotransfected with LMP1 or pSG5 plasmids, reporter plasmids of pGL3 vector control or EPHA4 promoter (−1000 ∼ +42) and internal control pEGFPC1 plasmids. Twenty-four hours posttransfection, 500 nM mithramycin was added to the cells for another 24 hours. EphA4-relative luciferase activity was determined as described previously (*P < .05, Student t test). Expression levels of LMP1 and β-actin were measured by western blotting (bottom panel). (F) BJAB cells were transduced with pSIN- or LMP1-expressing lentiviruses for 5 days and a chromatin immunoprecipitation assay was performed as described previously. DNA-protein complexes were immunoprecipitated using anti-Sp1 Ab or isotype control rabbit immunoglobulin G (IgG). EPHA4 promoter and control GAPDH promoter DNA were detected in the immunoprecipitates by PCR. Total DNA was harvested from BJAB cells and used as the input control.
The Eph JM domain, but not kinase domain, is involved in preventing LCL proliferation
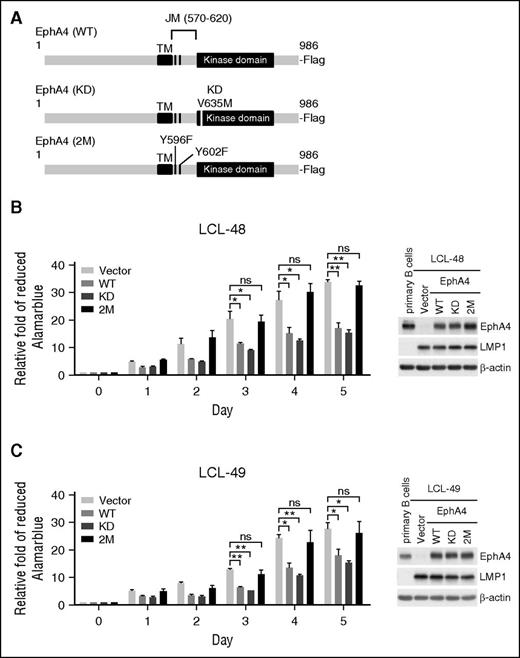
Like other EphA family members, EphA4 contains an extracellular domain, transmembrane (TM) and juxtamembrane (JM) domains, and an intracellular kinase domain.19 To understand the biological function of EphA4 in B cells, EphA4 full-length (WT) and mutants (including EphA4 with a kinase-dead mutant with V653M [KD] and EphA4 with mutations of the tyrosine autophosphorylation site Y569F and Y602F of the JM region (2M), as illustrated in Figure 5A), were delivered to LCLs by lentivirus infection. The transduced LCLs expressed WT or mutated EphA4 at the physiologic level, compared with primary B lymphocytes (Figure 5B-C right panel). Cell proliferation was inhibited significantly in the LCLs expressing EphA4 WT and KD but not in the LCLs expressing EphA4 2M (Figure 5B-C). These data showed that EphA4 repressed LCL proliferation through its JM region, suggesting that the kinase domain is not important for EphA4 repression of LCL proliferation.
The EphA4 JM domain is required for preventing LCL proliferation. (A) Flag-tagged EphA4 WT, KD (kinase dead with V653M), and 2M (JM region mutant with 2 tyrosine auto-phosphorylation sites Y569F and Y602F) expression plasmids for lentivirus packaging were illustrated. (B-C) LCLs from 2 donors were infected with EphA4 WT, mutants, or vector control-expressing lentiviruses for 2 days and then reseeded at 1 × 104 cells per well in 96-well plates for 5 days. Cell proliferation assays were measured by AlamarBlue reduction. Relative folds of proliferation were standardized with vector controls (*P < .05; **P < .01, Student t test). Total proteins were obtained from primary B cells and LCLs expressing WT and mutant forms of EphA4 at day 5 postreseeding. EphA4, LMP1, and β-actin expression levels were detected by western blotting (right panel). β-actin served as an internal control. ns, no significance.
The EphA4 JM domain is required for preventing LCL proliferation. (A) Flag-tagged EphA4 WT, KD (kinase dead with V653M), and 2M (JM region mutant with 2 tyrosine auto-phosphorylation sites Y569F and Y602F) expression plasmids for lentivirus packaging were illustrated. (B-C) LCLs from 2 donors were infected with EphA4 WT, mutants, or vector control-expressing lentiviruses for 2 days and then reseeded at 1 × 104 cells per well in 96-well plates for 5 days. Cell proliferation assays were measured by AlamarBlue reduction. Relative folds of proliferation were standardized with vector controls (*P < .05; **P < .01, Student t test). Total proteins were obtained from primary B cells and LCLs expressing WT and mutant forms of EphA4 at day 5 postreseeding. EphA4, LMP1, and β-actin expression levels were detected by western blotting (right panel). β-actin served as an internal control. ns, no significance.
EphA4 can be detected in tonsil biopsies but not in PTLD
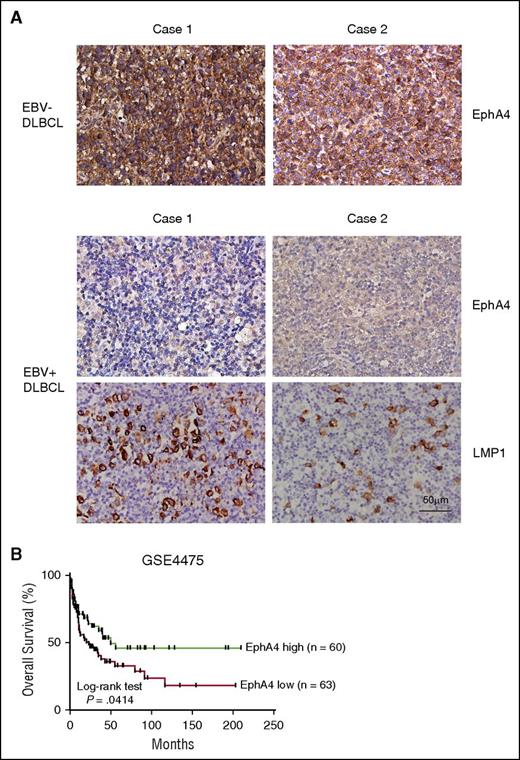
EBV+ PTLD exhibits similar biological features to LCLs. Thus, to address whether EphA4 was downregulated in PTLD biopsies, we examined EphA4 expression levels in 14 EBER+ PTLD biopsies and 5 EBER− tonsil biopsies by IHC assay. The results of the IHC assay showed that all cases of tonsil biopsies were positive for EphA4 staining. In addition, EphA4 was predominantly expressed in the cytoplasm and cell membranes in tonsil biopsies. On the other hand, 13 of 14 cases of PTLD (92.9%) were negative for EphA4 expression (Figure 6; Table 1). In addition, LMP1 was responsible for downregulation of EphA4 in our studies (Figures 2-4). Therefore, the expression levels of LMP1 were measured in EBER+ PTLD biopsies by IHC assay and 12 of 14 cases of PTLD (85.7%) were positive for LMP1 staining (Figure 6; Table 1).
Expression of EphA4 in PTLD biopsies. Paraffin-embedded PTLD and tonsils sections were subjected to IHC assays, and hematoxylin was used for the nuclear counterstaining. Positive signals of EphA4 were indicated as a brown color in tonsil biopsies but not in PTLD biopsies. LMP1 expression was also detected as a brown color by IHC assay in PTLD biopsies. The nuclei of the cells are colored blue. Magnification, ×200 (scale bar, 50 µm).
Expression of EphA4 in PTLD biopsies. Paraffin-embedded PTLD and tonsils sections were subjected to IHC assays, and hematoxylin was used for the nuclear counterstaining. Positive signals of EphA4 were indicated as a brown color in tonsil biopsies but not in PTLD biopsies. LMP1 expression was also detected as a brown color by IHC assay in PTLD biopsies. The nuclei of the cells are colored blue. Magnification, ×200 (scale bar, 50 µm).
EBV products and EphA4 in tissue biopsies
| . | Case . | EBER . | LMP1* . | EphA4 . | |||
|---|---|---|---|---|---|---|---|
| − . | + . | − . | + . | − . | + . | ||
| Tonsil | 5 | 5/5 | 0/5 | 5/5 | 0/5 | 0/5 | 5/5 |
| PTLD | 14 | 0/14 | 14/14 | 2/14 | 12/14 | 13/14 | 1/14 |
| DLBCL | 27 | 11/27 | 16/27 | 14/26* | 12/26* | 10/27 | 17/27 |
| . | Case . | EBER . | LMP1* . | EphA4 . | |||
|---|---|---|---|---|---|---|---|
| − . | + . | − . | + . | − . | + . | ||
| Tonsil | 5 | 5/5 | 0/5 | 5/5 | 0/5 | 0/5 | 5/5 |
| PTLD | 14 | 0/14 | 14/14 | 2/14 | 12/14 | 13/14 | 1/14 |
| DLBCL | 27 | 11/27 | 16/27 | 14/26* | 12/26* | 10/27 | 17/27 |
One case of LMP1 status was not determined in EBER+ DLBCL.
A reverse correlation between EphA4 and the survival rate of EBV+ DLBCL is revealed
According to 2016 World Health Organization (WHO) classification, EBV+ DLBCL of the elderly, which is equivalent to EBV+ DLBCL, not otherwise specified, displays a similar EBV viral protein expression as LCL.2,34 To confirm that EphA4 expression is correlated with EBV infection and LMP1 expression, 16 EBER+ DLBCL and 11 EBER− DLBCL biopsies from patients without HIV infection or any iatrogenic immunosuppression were examined. The IHC assay indicated that all cases of EBV− DLBCL biopsies were positive for EphA4 staining (Figure 7A; Tables 1 and 2). In contrast, 10 of 16 cases of EBV+ DLBCL (62.5%) were negative for EphA4 expression, 3 showed very weak, positive signals, and 3 were stained positively (Figure 7A; Tables 1 and 2). That the presence of EBV was associated with reduced EphA4 expression was statistically significant (Table 2, P = .0011, Fisher exact test). On the other hand, 12 of 15 cases of EBV+ DLBCL (80%) were positive for LMP1 staining (Figure 7A; Table 1). LMP1 expression was also inversely correlated with EphA4 expression (Table 2, P = .0375, Fisher exact test). Next, we asked whether there was correlation between EphA4 expression and patients’ survival. So, we analyzed the survival data of patients with DLBCL from GEO data sets (accession number GSE4475). The profile of overall survival in patients was used to estimate whether patients with low EphA4 expression have worse prognosis. Based on the median expression value of EphA4, 123 patients were grouped into 2 clusters; 60 patients had high EphA4 and 63 patients had low EphA4. Patients with low EphA4 had worse overall survival (Figure 7B, P = .0414). Our results indicated that low EphA4 expression may potentially contribute to a poor prognosis for these DLBCL patients.
Detection of EphA4 expression in DLBCL biopsies. (A) Paraffin-embedded DLBCL sections were stained for EphA4. Positive signals of EphA4 could be seen as a brown color in EBV− DLBCL, but not in EBV+ DLBCL biopsies, by IHC. LMP1 expression was also detected in EBV+ DLBCL biopsies. The nuclei were observed as a blue color and hematoxylin was used for the nuclear counterstain. Magnification, ×200 (scale bar, 50 µm). (B) This survival curve of DLBCL was obtained from the GEO data sets. Patients were divided into high (n = 60) and low EphA4 (n = 63) groups, according to the median expression level of EphA4. A Kaplan-Meier plot showed that patients with low EphA4 had worse overall survival (P = .0414, log-rank test).
Detection of EphA4 expression in DLBCL biopsies. (A) Paraffin-embedded DLBCL sections were stained for EphA4. Positive signals of EphA4 could be seen as a brown color in EBV− DLBCL, but not in EBV+ DLBCL biopsies, by IHC. LMP1 expression was also detected in EBV+ DLBCL biopsies. The nuclei were observed as a blue color and hematoxylin was used for the nuclear counterstain. Magnification, ×200 (scale bar, 50 µm). (B) This survival curve of DLBCL was obtained from the GEO data sets. Patients were divided into high (n = 60) and low EphA4 (n = 63) groups, according to the median expression level of EphA4. A Kaplan-Meier plot showed that patients with low EphA4 had worse overall survival (P = .0414, log-rank test).
Discussion
Physiologically, Eph receptors and their ligands are critical for many developmental processes. Pathologically, the Eph family is dysregulated in a variety of human diseases, especially cancers. During cancer formation, Eph can function in tumor suppression or tumor promotion, depending on the type of cancer, the interacting ligands and cross-talk with other RTKs. Among them, EphA2 is a good example. Limited expression of EphA2 is required for mammary gland development but its overexpression may enhance the malignancy of breast cancer.35 In brain tumors, the presence of ephrin-A1 determines the direction of a reciprocal loop between EphA2 and Akt, which results in inhibition or promotion of the tumor grade.36 In addition to neuronal development, EphA4 has been found to be involved in tumor progression.18,19,21,22,37,38 For example, EphA4 is upregulated in colon cancer with liver metastasis.22 In invasive cervical carcinoma, EphA4 expression is decreased due to chromosomal deletions, with loss of heterozygosity.21 EphA4 mRNA is downregulated or lost in metastatic melanoma.37 The regulatory mechanisms of EphA4 mRNA expression have been explored in terms of transcription. In the cervical cancer cell line HeLa and U373 glioma cells, EphA4 is reduced due to mRNA instability through regulation of its 3′ untranslated regions by HuR.39 In hepatocellular carcinoma, microRNA-10a targets EphA4 mRNA and overexpression of EphA4 inhibits cell migration and invasion.40 Moreover, a Pax3/FKHR oncogenic fusion protein has been shown to bind directly to the promoter region of EphA4, thus increasing EphA4 transcription in SaOS-2 cells.41 Furthermore, DNA methylation of EphA4 is observed in acute lymphoblastic leukemia.42 Our results showed that EBV LMP1 inhibits EphA4 promoter activity through Sp1 (Figure 4).
Sp1, which is expressed ubiquitously, can serve as a transactivator or repressor to regulate the promoter activities of target genes via phosphorylation.43 For example, FGF-2 stimulation enhances Sp1 binding to repress the promoter of platelet-derived growth factor receptor-α in smooth muscle cells through ERK-mediated phosphorylation of Sp1 at T453 and T739.44 HER2 also has been reported to suppress the RECK promoter by ERK-mediated Sp1phosphorylation.45 In this study, LMP1 activated the ERK pathway to inhibit the EphA4 promoter through Sp1 binding (Figure 4). We provide this new insight into how a viral oncoprotein can regulate Eph expression.
The function of EphA4 associated with cancer progression is controversial. As a tumor suppressor, EphA4 downregulates ERK phosphorylation to inhibit migration and invasion in non-small cell lung cancer cells.38 However, EphA4 can act as tumor promoter; for example, EphA4 interacts directly with FGFR1 through its cytoplasmic domains, phosphorylates the docking protein, FRS2α, and enhances cell migration and proliferation.46,47 In our case, EphA4 prevents LCL from proliferation (Figure 5), suggesting that it probably plays an inhibitory role in EBV+ B-lymphoproliferative disorders. This is confirmed by the IHC results of PTLD and DLBCL (Figures 6A-7A). In this study, expression of EphA4 was undetectable in the cells of EBER+ PTLD biopsies (Figure 6; Table 1). Because the incidence of EBV in PTLD cases is over 90%,2 these data imply that EphA4 is involved in the pathogenesis of EBV+ PTLD.
In addition to PTLD, our results indicated that cells in EBV+ DLBCL biopsies exhibited lower expression levels of EphA4 than those in EBV− biopsies (Figure 7A; Table 2), verifying the inverse correlation of EphA4 and EBV infection in DLBCL patients. It has been reported that patients with EBV+ DLBCL have worse overall survival and progression-free survival than their EBV− counterparts,34,48 yet the underlying mechanism has not been fully understood. Here, we provided observation to show that low EphA4 correlated with poor survival outcome for DLBCL (Figure 7B). Also, some EphA4 ligands, such as ephrin-A1, A2, A3, A4, and B1 were downregulated post-EBV infection (data not shown). The results of overexpression of EphA4 in LCL indicated that EphA4 prevention of B-cell proliferation may be ligand-independent.
Taken together, LMP1-mediated EphA4 repression accelerates B-cell proliferation post-EBV infection. Our clinical results suggest that downregulation of EphA4 in patients with PTLD and DLBCL may provide new insights into pathogenesis and a poor prognostic marker for EBV-associated B-cell malignancies. EphA4 may be considered as a potential therapy target for PTLD and DLBCL.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Tim J. Harrison of University College London Medical School (London, United Kingdom) for reviewing the manuscript critically. The authors thank Taipei Blood Center of Taiwan Blood Service Foundation for providing whole blood.
This work was supported by the Ministry of Science and Technology (grant: 103-2320-B-002-038-MY3), the National Health Research Institute (grant: NHRI-EX105-10306BI), Excellent Translational Medicine Research Projects of National Taiwan University College of Medicine and National Taiwan University Hospital (grant: 105R39012) (C.-H.T.), and by the Ministry of Science and Technology (grant: 103-2320-B-182-028-MY3) (S.-J.L.).
Authorship
Contribution: Y.-C.H. designed experiments, performed experiments, analyzed the data, and cowrote the manuscript; S.-J.L. and C.-H.T. designed experiments and cowrote the manuscript; K.-M.L. and J.L. performed experiments; Y.-C.C. performed experiments and analyzed the data; C.-W.L., T.-L.S., and M.-R.C. provided materials; and S.-C.Y., C.-L.C., and C.-K.C. provided materials and analyzed the data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ching-Hwa Tsai, Graduate Institute of Microbiology, College of Medicine, National Taiwan University, Room 719, No. 1, 1st Section, Jen-Ai Rd, Taipei 10051, Taiwan; e-mail: chtsai@ntu.edu.tw.

![Figure 1. EphA4 expression is decreased post-EBV infection. (A) Total RNAs were harvested from uninfected primary B cells, from peripheral blood, and EBV-immortalized LCLs from 3 different donors. cDNAs were generated using [33P]-labeled degenerate primers for PTKs. EphA4-specific cDNAs were digested by 3 restriction enzymes, BstNI, HhaI, and MnlI. Relative fold EphA4 expression was compared with primary B cells. (B-C) Peripheral CD19+ B cells were seeded at 1 × 106 cells/mL and infected with EBV strain B95.8. Total RNAs and protein were harvested from primary B cells at the days indicated post-EBV infection. (B) Expression levels of EphA4 mRNA were measured by RT-Q-PCR. EphA4 mRNA-relative folds were normalized to internal control MAGOH and standardized with uninfected primary B cells. (C) EphA4, EBNA1, LMP1, and β-actin proteins were measured by western blotting. EphA4 protein-relative folds were normalized to internal control β-actin and compared with uninfected primary B cells. (D) Total RNAs were extracted from primary B cells, EBV infection, or B-cell stimulations including anti-CD40 antibody plus IL-4, LPS, or poly I:C for 3 days. EphA4 transcripts were measured by RT-Q-PCR. EphA4 mRNA-relative folds were normalized to internal control MAGOH and standardized with uninfected primary B cells. (E) Total RNAs were extracted from primary B cells and 8 LCL lines. Expression levels of EphA4 mRNA were measured by RT-Q-PCR. EphA4 mRNA-relative folds were normalized to internal control MAGOH and standardized with uninfected primary B cells. (F-G) Total RNAs and protein were harvested from paired uninfected B cells and LCLs generated from the peripheral blood mononuclear cells of 2 healthy donors. (F) Expression levels of EphA4 mRNA were measured by RT-Q-PCR. EphA4 mRNA-relative folds were normalized to internal control MAGOH and standardized with uninfected primary B cells. (G) EphA4, EBNA1, LMP1, and β-actin proteins were measured by western blotting. EphA4 protein-relative folds were normalized to β-actin and compared with uninfected primary B cells.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/128/12/10.1182_blood-2016-02-702530/4/m_1578f1.jpeg?Expires=1765899933&Signature=OA6cZXDap-AP6m5A5GEnDeF169ESRu0kqG75dWJscl21ApNIb4qF2FbHsBtEJnS~5~2JmgloMxhls-glh86EUB60atZE5fx7CJMcbog0Qim6THFCCGc76vTEIsnVT-PowZherEK24WJmLVXa6VOhg88Tb0OpSaMSIlhZyL4eycpscNgS5YAGcOY-7CmZEicxfZhHiQVRtO0-oS9WmegnbIDKv34j3yn4RsR8kbvoDsFx4vl5v4e3pWvSoLcUrBN-Z2b7qG1dFZIzplwRbqxwtO8LwUFrW1b62XydGa6mr57n4rw61M~BE3Y1A3vOcqkR~lPbYOPvqKIP5o0i2plkug__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal