Key Points
Germ line loss-of-function mutations in shelterin genes occur in a subset of families with CLL.
Telomere dysregulation is further implicated in CLL predisposition.
Abstract
Chronic lymphocytic leukemia (CLL) can be familial; however, thus far no rare germ line disruptive alleles for CLL have been identified. We performed whole-exome sequencing of 66 CLL families, identifying 4 families where loss-of-function mutations in protection of telomeres 1 (POT1) co-segregated with CLL. The p.Tyr36Cys mutation is predicted to disrupt the interaction between POT1 and the telomeric overhang. The c.1164-1G>A splice-site, p.Gln358SerfsTer13 frameshift, and p.Gln376Arg missense mutations are likely to impact the interaction between POT1 and adrenocortical dysplasia homolog (ACD), which is a part of the telomere-capping shelterin complex. We also identified mutations in ACD (c.752-2A>C) and another shelterin component, telomeric repeat binding factor 2, interacting protein (p.Ala104Pro and p.Arg133Gln), in 3 CLL families. In a complementary analysis of 1083 cases and 5854 controls, the POT1 p.Gln376Arg variant, which has a global minor allele frequency of 0.0005, conferred a 3.61-fold increased risk of CLL (P = .009). This study further highlights telomere dysregulation as a key process in CLL development.
Introduction
Chronic lymphocytic leukemia (CLL; MIM151400) is clinically defined by the presence of a clonal population of B-cell lymphocytes (>5 × 109 cells/L) with a characteristic immunophenotype. The disease accounts for ∼25% of all leukemia and is the most common form of lymphoid malignancy in Western countries, affecting ∼16 000 individuals in the United States each year.1 Although the last decade has seen a dramatic evolution in the treatment options for CLL,2-4 it still remains an incurable malignancy. It is anticipated that an increased understanding of CLL pathogenesis will generate further therapeutic targets to either delay or prevent progression of the precursor to frank malignancy.
CLL has one of the highest familial risks of any cancer, with risk being increased eightfold in relatives of patients.5 Recent genome-wide association studies (GWAS) have identified common risk single nucleotide polymorphisms (SNPs) at 31 loci associated with sporadic CLL.6-13 The risk of CLL associated with each of these variants is however modest at best. Although families segregating CLL provide evidence for Mendelian susceptibility, no rare alleles of large effect have thus far been discovered. The identification of this class of susceptibility is especially important because mutations are causal and provide direct insight to cancer biology, in contrast to GWAS associations.
Here we report on the whole exome sequencing (WES) of familial CLL, and establish a key role for rare disruptive mutations in protection of telomeres 1 (POT1) and other shelterin complex genes as determinants of susceptibility to CLL. Our findings thus extend the spectrum of cancer types associated with germ line mutation in these genes.
Materials and methods
Patient samples and DNA extraction
The families and CLL cases included in this study were recruited through a United Kingdom national study of CLL genetics, established by The Institute of Cancer Research (ICR) Divisions of Genetics and Epidemiology and Molecular Pathology in 1996. The diagnosis of CLL and other hematologic cancers in family members were established. In all cases, the diagnosis of CLL was based on accepted standard clinico-pathological and immunologic criteria that are in accordance with current World Health Organization classification guidelines. Informed consent was obtained under the Multi-Research Ethics Committee 99/1/082. Genomic DNA was extracted from peripheral blood and saliva using standard methods, and quantified by PicoGreen (Invitrogen).
Pedigrees and clinical presentation
See supplemental Table 1, available on the Blood Web site, for details on all pedigrees, the number of cases of CLL in each family, and the number of cases that were whole-exome sequenced.
Sequence alignment and analysis
Exon capture was performed using the Nextera Rapid Capture Exome Enrichment Kit (Illumina, San Diego, CA). The Illumina HiSeq 2000 analyzer with 101 bp reads was used for sequencing. Paired-end FASTQ files were extracted using CASAVA software (version 1.8.1; Illumina) and aligned to build 37 (hg19) of the human reference genome using Stampy14 and Burrows-Wheeler Aligner15 software. Alignments were processed using the Genome Analysis Tool Kit pipeline (version 3.2-2),16 according to best practices.17,18 Variants were filtered for positions found in >1 sample from an in-house collection of 1609 control exomes, including 961 samples from the ICR1000 data set generated by Nazneen Rahman’s team in the Division of Genetics and Epidemiology at the ICR, London, United Kingdom,19 plus an extra 648 samples from the UK 1958 Birth Cohort (BC),20 sequenced in-house using Illumina TruSeq exome methodology. We also filtered variants based on frequencies in the 1000 Genomes Project, National Heart, Lung, and Blood Institute Exome Sequencing Project (ESP6500), and the Exome Aggregation Consortium (ExAC) catalog. Positions resulting in protein-altering changes were identified using the Ensembl Variant Effect Predictor (version 78) and variants shared between family members were annotated using custom scripts. The predicted functional consequences of missense variants were assessed using SIFT,21 CADD,22 and SuSPect23 algorithms.
Sanger sequencing
Germ line verification of variants found by next-generation sequencing was performed by Sanger sequencing of mouthwash DNA samples. Primers are listed in supplemental Table 2.
MaxEntScan scoring of splice acceptor variants
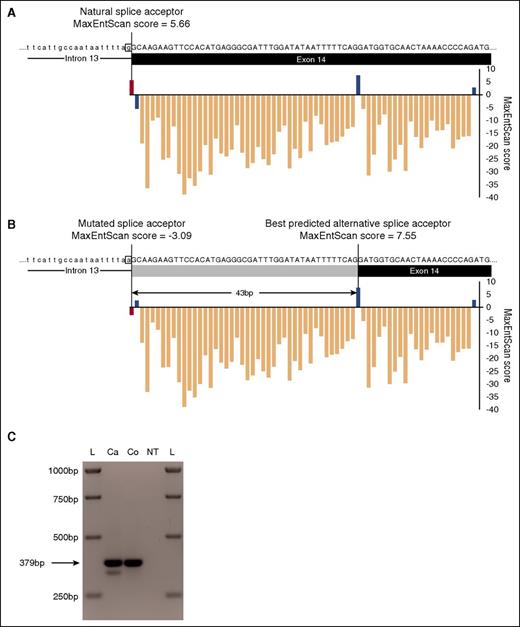
We used the MaxEntScan algorithm24 to assess the effect of POT1 g.124481233C>T and adrenocortical dysplasia homolog (ACD) g.67692984T>G mutations. Scores for the mutated splice acceptor site and wild-type (WT) splice site sequence were 5.66; −3.08 and 9.88, and 1.84, respectively.
Confirmation of aberrant splicing in an individual carrying the splice acceptor variant, 7:g.124481233C>T
RNA extracted from the whole blood of a splice acceptor variant carrier and control was converted to complementary DNA (cDNA) using Superscript III Reverse Transcriptase (Invitrogen). Polymerase chain reaction (PCR) was then performed to confirm that 7:g.124481233C>T disrupted splicing. The product was visualized on a 2.5% agarose gel. Sanger sequencing was used to confirm the sequence of the product.
Exome array genotyping
A total of 1111 unrelated CLL cases were genotyped for the p.Gln376Arg variant using the Illumina OmniExpress Exome array as previously described.12 After quality control filtering, genotype data were available for 1083 CLL cases. For controls, we used publicly accessible data for 5854 individuals from the 1958 BC,20 genotyped using the Illumina HumanExome-12 version 1 array. These data are available from the European Genome-phenome Archive under accession #EGAD00010000234. The χ2 test was used to determine the significance of the difference in case-control allele counts. Confidence intervals (CIs) were calculated by the Woolf method.
Protein alignment and structural modeling
Multiple sequence alignments were generated for homologous POT1 and telomeric repeat binding factor 2, interacting protein (TERF2IP) sequences using T-Coffee25,26 to evaluate conservation. POT1 alignments were generated with the following sequences: NP_056265.2, XP_519345.2, NP_001127526.1, XP_009001386.1, XP_006149256.1, NP_598692.1, XP_002712135.2, XP_010802750.1, XP_005628494.1, XP_001501458.4, XP_006910616.1, XP_010585693.1, XP_004478311.1, XP_007504310.1, XP_001508179.2, NP_996875.1, and NP_001084422.1. TERF2IP alignments were generated with the following sequences: NP_061848.2, NP_001267142.1, XP_003780774.2, XP_008984478.1, XP_006152679.1, NP_065609.2, XP_002711780.1, NP_001068880.1, XP_536776.2, XP_005608497.1, XP_006908867.1, XP_010595146.1, XP_004470975.1, XP_001508762.2, NP_989799.1, and NP_001084428.1. Jalview27 was used to visualize and format the alignments. The crystal structure of the N-terminal region (oligonucleotide/oligosaccharide-binding 1 [OB1] and OB2 domains) of the human POT1 protein (Research Collaboratory for Structural Bioinformatics protein data bank [PDB], 3KJP, and 1XJV) was visualized using Chimera (version 1.10.2)28 and Cn3D (version 4.3.1).29 The impact of the p.Tyr36Cys mutation on stability of the POT1:DNA interaction was assessed using the mutation Cutoff Scanning Matrix (mCSM) approach.30 The effect of missense mutations on protein stability was assessed using the Impact of Non-synonymous mutations on Protein Stability (INPS) server.31
LOH analyses
Loss-of-heterozygosity (LOH) analysis was conducted using ExomeCNV,32 which detects copy number variation and LOH events using depth-of-coverage and B-allele frequencies. LOH calls were made by first identifying all heterozygous germ line positions. The Genome Analysis Tool Kit was then used to create BAF files and ExomeCNV was used to call LOH at heterozygous positions individually and at combined LOH segments.
Assessment of telomere length
Relative telomere length was determined by 2 methods: using exome sequencing data and with real-time PCR (RT-PCR). Analysis of off-target reads from exome sequencing data were performed essentially as described,33 using a telomeric repeat copy number of k = 4. We used data from blood-derived DNA only and also excluded samples with average sequencing depth <20 and with missing covariate data (n = 12). Telomere length was adjusted for age at blood draw, sex, and sequencing batch by a linear model determined using data from noncarriers only. For the SYBR green RT-PCR, the ratio of telomere repeat units to a single-copy gene (β-globin) for 109 samples, was determined as previously described.34,35 Primers are listed in supplemental Table 2. Reactions were performed in triplicate, using 10 ng DNA per sample. Each 10 μL reaction also contained 5 μL of 2X SYBR Green Master Mix (Applied Biosystems), plus either 300 and 700 nmol/L of the control forward and reverse primers, respectively, or 100 and 900 nmol/L of the telomere unit forward and reverse primers, respectively. The telomere reaction also included 0.3 μL dimethyl sulfoxide. Cycling was performed using an ABI7900HT thermal cycler as previously described.35 Relative telomere length was calculated using 2−ΔCt derived from the RT-PCR data and was adjusted for age at blood draw and sex. A Wilcoxon rank-sum test was used to compare the relative adjusted telomere length for POT1 mutation carriers vs noncarriers of shelterin gene mutations.
Results
Identification of shelterin gene mutations
To maximize the prospects of identifying rare disease-causing variants for CLL, we initially focused our search on the 18 families with the strongest family histories of CLL (supplemental Table 1), which had been ascertained through an ongoing study.36 We performed WES on genomic DNA from blood of 45 affected individuals from the 18 families. We excluded variants that were observed more than once in our in-house database of 1609 healthy individuals from the 1958 BC who had been exome sequenced. We also discounted variants with an allele frequency of >0.1% in large-scale sequencing projects (1000 Genomes Project, ESP6500, or the ExAC catalog). In our first stage analysis, we required the filtered variants to be present in all sequenced affecteds within the family.
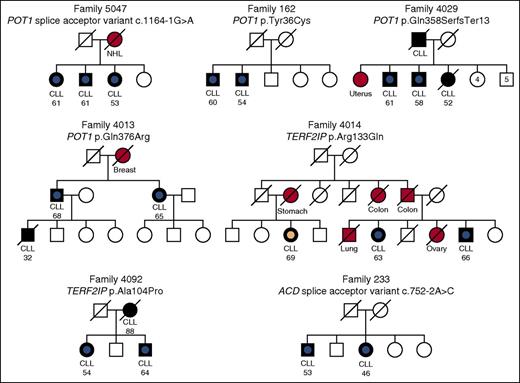
To further filter the variants identified, we prioritized missense and disruptive variants (nonsense, splice acceptor/donor, and frameshift) occurring in genes with a reported cancer association or documented role in cancer predisposition. Analysis of these genes led us to identify pedigree 5047, in which all 3 affected family members carried a splice acceptor variant in intron 13 (chromosome 7 g.124481233C>T/c.1164-1G>A) of POT1 (MIM 606478) (Figures 1 and 2A).37-40 We confirmed the mutation by Sanger sequencing in blood and saliva-derived DNA in all 3 cases (supplemental Figure 1). The mutation was predicted to disrupt splicing by the MaxEntScan algorithm24 (WT score 5.66 vs mutated score −3.08; 154% reduction). This also identified a potential alternative splice acceptor site 43 bp downstream (MaxEntScan score = 7.66), the use of which would result in a truncated protein product. We confirmed the presence of an aberrant splicing product in a mutation carrier by RT-PCR and validated the use of the predicted alternative splice site using Sanger sequencing (Figure 3 and supplemental Figure 2).
Rare POT1, ACD, and TERF2IP mutations in CLL families. Black-filled symbols indicate CLL cases, other cancers are indicated by a red-filled symbol, and an unfilled symbol indicates an individual with no known cancer. These symbols have a central dot to indicate cases that were exome sequenced. A central blue dot denotes a shelterin gene mutation carrier; a peach dot denotes a WT individual. A line through a symbol indicates that an individual is deceased. Age of diagnosis (in years) is listed for CLL cases. Splice acceptor variants are numbered relative to POT1 transcript NM_015450 and ACD transcript NM_001082486. NHL, non-Hodgkin lymphoma.
Rare POT1, ACD, and TERF2IP mutations in CLL families. Black-filled symbols indicate CLL cases, other cancers are indicated by a red-filled symbol, and an unfilled symbol indicates an individual with no known cancer. These symbols have a central dot to indicate cases that were exome sequenced. A central blue dot denotes a shelterin gene mutation carrier; a peach dot denotes a WT individual. A line through a symbol indicates that an individual is deceased. Age of diagnosis (in years) is listed for CLL cases. Splice acceptor variants are numbered relative to POT1 transcript NM_015450 and ACD transcript NM_001082486. NHL, non-Hodgkin lymphoma.
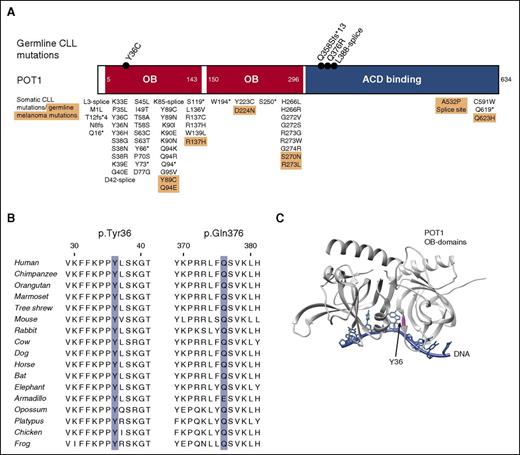
Impact of rare familial mutations on POT1 protein. (A) Schematic showing the position of germ line POT1 mutations identified in CLL families relative to OB domains (red) and ACD binding region (blue). Also shown are somatic POT1 mutations identified in previous studies of CLL patients37,38 (unshaded background) and germ line mutations found in familial cutaneous melanoma39,40 (peach background). (B) Cross-species conservation of POT1 amino acids subject to missense mutation in CLL families. (C) Schematic of the crystal structure of human POT1 N-terminal OB domains bound to a telomeric DNA sequence (PDB 3KJP), illustrating the proximity of tyrosine 36 to the DNA strand. OB domains are shown in gray, DNA in blue, and Tyr.36 is highlighted in magenta.
Impact of rare familial mutations on POT1 protein. (A) Schematic showing the position of germ line POT1 mutations identified in CLL families relative to OB domains (red) and ACD binding region (blue). Also shown are somatic POT1 mutations identified in previous studies of CLL patients37,38 (unshaded background) and germ line mutations found in familial cutaneous melanoma39,40 (peach background). (B) Cross-species conservation of POT1 amino acids subject to missense mutation in CLL families. (C) Schematic of the crystal structure of human POT1 N-terminal OB domains bound to a telomeric DNA sequence (PDB 3KJP), illustrating the proximity of tyrosine 36 to the DNA strand. OB domains are shown in gray, DNA in blue, and Tyr.36 is highlighted in magenta.
Impact of POT1 splice acceptor site mutation on splicing. (A) Splice acceptor site consensus scores predicted by MaxEntScan24 for each base from the natural POT1 intron 13/exon 14 boundary across exon 14. For clarity, only part of the intron (lower case text above black line) and exon (upper case text above black box) are shown. The predicted score for the unmutated natural splice site (red bar) is also labeled. Positive scores are otherwise marked in blue and negative scores in peach. (B) MaxEntScan splice acceptor consensus scores for the same region based upon the sequence of c.1164-1G>A POT1 mutation carriers. The scores of the mutated natural splice acceptor (pink bar) and the predicted alternative splice site with the highest MaxEntScan score (43 bp downstream) are labeled. The part of exon 14 that would be removed by use of this splice site is indicated by a gray box. (C) Abnormal splicing product detected by RT-PCR using cDNA from a CLL case (Ca) carrying the c.1164-1G>A mutation. This product is absent from control (Co) cDNA. bp, base pairs; L, ladder; NT, no template reaction.
Impact of POT1 splice acceptor site mutation on splicing. (A) Splice acceptor site consensus scores predicted by MaxEntScan24 for each base from the natural POT1 intron 13/exon 14 boundary across exon 14. For clarity, only part of the intron (lower case text above black line) and exon (upper case text above black box) are shown. The predicted score for the unmutated natural splice site (red bar) is also labeled. Positive scores are otherwise marked in blue and negative scores in peach. (B) MaxEntScan splice acceptor consensus scores for the same region based upon the sequence of c.1164-1G>A POT1 mutation carriers. The scores of the mutated natural splice acceptor (pink bar) and the predicted alternative splice site with the highest MaxEntScan score (43 bp downstream) are labeled. The part of exon 14 that would be removed by use of this splice site is indicated by a gray box. (C) Abnormal splicing product detected by RT-PCR using cDNA from a CLL case (Ca) carrying the c.1164-1G>A mutation. This product is absent from control (Co) cDNA. bp, base pairs; L, ladder; NT, no template reaction.
To further investigate the potential role of POT1 and other members of the shelterin gene complex in familial CLL, we expanded our exome sequencing data set to include an additional 96 affected relative-pairs from 48 families (supplemental Table 1). We then looked for shared missense and disruptive variants in the 6 components of the shelterin complex (POT1, ACD, telomeric repeat binding factor 1 [TERF1] interacting nuclear factor 2 [TINF2], TERF1, TERF2, and TERF2IP). Through this analysis, we identified 3 additional families that harbored POT1 mutations (Figure 1); 2 missense mutations, p.Tyr36Cys and p.Gln376Arg, occurring at evolutionarily conserved residues (Figure 2B), predicted in silico to be damaging by multiple algorithms, and a frameshift mutation (Table 1). Collectively, we therefore identified mutations in POT1 in 6% of the CLL families, as compared with the documented frequency of such variants of only 0.9% among the 60 706 individuals included in the ExAC catalog (P = .003).
Germ line mutations in shelterin complex genes identified in CLL pedigrees
| Gene . | Mutation position . | . | . | . | Effect predictions . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genomic (hg19) . | cDNA* . | Protein . | Variant type . | Pedigree . | Carriers† . | CADD‡ . | SIFT . | SuSPect§ . | GERP . | |
| POT1 | 7:g.124481233C>T | c.1164-1G>A | N/A | Splice acceptor | 5047 | 3/3/3 | 17.16 | N/A | N/A | 4.67 |
| POT1 | 7:g.124532337T>C | c.107A>G | p.Tyr36Cys | Missense | 162 | 2/2/2 | 18.9 | Deleterious | 71 | 5.71 |
| POT1 | 7:g.124482952_124482953insA | c.1071_1072insT | p.Gln358SerfsTer13 | Frameshift | 4029 | 2/2/4 | N/A | N/A | N/A | 4.71 |
| POT1 | 7:g.124482897T>C | c.1127A>G | p.Gln376Arg | Missense | 4013 | 2/2/3 | 23 | Deleterious | 50 | 5.55 |
| ACD | 16:g.67692984T>G | c.752-2A>C | N/A | Splice acceptor | 233 | 2/2/2 | 19.35 | N/A | N/A | 5.15 |
| TERF2IP | 16:g.75682090G>C | c.310G>C | p.Ala104Pro | Missense | 4092 | 2/2/3 | 10.15 | Tolerated | 10 | 2 |
| TERF2IP | 16:g.75682178G>A | c.398G>A | p.Arg133Gln | Missense | 4014 | 2/3/3 | 14.88 | Deleterious | 73 | 5.34 |
| Gene . | Mutation position . | . | . | . | Effect predictions . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genomic (hg19) . | cDNA* . | Protein . | Variant type . | Pedigree . | Carriers† . | CADD‡ . | SIFT . | SuSPect§ . | GERP . | |
| POT1 | 7:g.124481233C>T | c.1164-1G>A | N/A | Splice acceptor | 5047 | 3/3/3 | 17.16 | N/A | N/A | 4.67 |
| POT1 | 7:g.124532337T>C | c.107A>G | p.Tyr36Cys | Missense | 162 | 2/2/2 | 18.9 | Deleterious | 71 | 5.71 |
| POT1 | 7:g.124482952_124482953insA | c.1071_1072insT | p.Gln358SerfsTer13 | Frameshift | 4029 | 2/2/4 | N/A | N/A | N/A | 4.71 |
| POT1 | 7:g.124482897T>C | c.1127A>G | p.Gln376Arg | Missense | 4013 | 2/2/3 | 23 | Deleterious | 50 | 5.55 |
| ACD | 16:g.67692984T>G | c.752-2A>C | N/A | Splice acceptor | 233 | 2/2/2 | 19.35 | N/A | N/A | 5.15 |
| TERF2IP | 16:g.75682090G>C | c.310G>C | p.Ala104Pro | Missense | 4092 | 2/2/3 | 10.15 | Tolerated | 10 | 2 |
| TERF2IP | 16:g.75682178G>A | c.398G>A | p.Arg133Gln | Missense | 4014 | 2/3/3 | 14.88 | Deleterious | 73 | 5.34 |
GERP, Genomic Evolutionary Rate Profiling score; N/A, not applicable.
POT1 reference transcript is NM_015450 and ACD reference transcript is NM_001082486.
Carriers given as number of familial cases with mutation/number of cases in family exome sequenced/total number of CLL cases in family.
CADD Phred-like score.
Scores of 50 and above considered to indicate deleterious mutations.
Intriguingly, somatic mutations of residue Tyr36 have previously been reported in CLL (Figure 2A).37,41,42 The p.Gln376Arg variant, identified in pedigree 4013, has a global minor allele frequency of 0.0005 in the ESP6500 database and is included on the Illumina Exome array. We therefore initiated a genetic association study of this recurrent variant, making use of Illumina exome array data on 1083 unselected CLL cases and 5854 1958 BC controls. Six of the cases and 9 of the controls were heterozygous for the p.Gln376Arg variant (odds ratio = 3.61; 95% CI, 1.28-10.15; P = .009).
In addition to POT1 mutations, we identified mutations in other shelterin complex genes in families 233, 4092, and 4014. Specifically, the ACD (MIM 609377) splice site variant c.752-2A>C was carried by both affected siblings in pedigree 233 (Figure 1 and supplemental Figure 3A) and was predicted by the MaxEntScan algorithm to disrupt the exon 7 splice acceptor signal (WT score 9.88 vs mutated score 1.84; 81% reduction) (supplemental Figure 3B). In TERF2IP (MIM 605061), the missense mutation c.398G>A (p.Arg133Gln) was identified in 2 out of 3 CLL cases sequenced in family 4014 (Figure 1 and supplemental Figure 4A). This mutation occurs at an evolutionarily conserved site and was predicted to be damaging by multiple methods (Table 1; supplemental Figure 4B). We also found the c.310G>C (p.Ala104Pro) TERF2IP variant in both siblings in family 4092 (Figure 1; Table 1; supplemental Figure 4A). Although this residue is partially conserved, the p.Ala104Pro mutation is not predicted to be damaging by SIFT or SuSPect (Table 1; supplemental Figure 4B).
Structural predictions
The POT1 N-terminus contains 2 OB folds that bind to the single-stranded telomeric overhang (Figure 2), whereas the C-terminus is responsible for binding to ACD and anchoring the shelterin complex. The crystal structure of human POT1 has been resolved for only the N-terminal OB folds (PDB 3KJP and 1XJV). Based upon these structures, Tyr36 is one of 24 residues found at the POT1:telomeric polynucleotide interface37 (Figure 2). The p.Tyr36Cys mutation is predicted by the mutation Cutoff Scanning Matrix approach to reduce the POT1:DNA complex affinity (PDB 1XJV, ΔΔG −0.27 kcal/mol; PDB 3KJP, predicted ΔΔG −0.21 kcal/mol).
Because crystal structures for full-length POT1 and TERF2IP are lacking, we used the machine learning algorithm Impact of Non-synonymous mutations on Protein Stability to predict the thermodynamic change in free energy caused by the p.Gln376Arg (POT1), p.Ala104Pro, and p.Arg133Gln (TERF2IP) mutations, based upon the protein sequence (supplemental Table 3). Using this method, p.Arg133Gln was predicted to have the largest effect upon protein stability.
Analysis of somatic events
We used ExomeCNV to look for evidence of LOH in the proband of pedigree 5047, comparing exome sequencing data from blood-derived DNA to saliva-derived DNA, finding no evidence of a somatic abnormality at the POT1 locus. We also looked for deleterious variants identified only in the blood-derived DNA of this case (ie, absent from the saliva-derived DNA sample and also absent from the other affected individuals in pedigree 5047), and found no somatic inactivating POT1 mutations. We did however, note the presence of a somatic splice donor site mutation affecting the first base of intron 10 of ATR (or ataxia-telangiectasia and rad3-related) in this case.
Effect of POT1 mutations on maintenance of telomere length
Given the role of the shelterin complex in telomere length maintenance, we examined whether CLL cases from shelterin-mutated pedigrees had telomere lengths that differed from noncarrier CLL cases using exome sequencing and RT-PCR data. We observed no consistent significant difference between the telomere lengths of POT1 mutation carriers and CLL cases without a mutation in a shelterin complex gene, by exome sequencing or RT-PCR (P = .03 and P = .57, respectively). The telomere lengths of cases with ACD or TERF2IP variants also displayed no obvious trend, although the small numbers of cases harboring these variants precluded a meaningful evaluation of their impact on telomere length.
Discussion
Here we have implemented WES to search for rare disruptive risk alleles for CLL, identifying germ line-inactivating shelterin gene mutations in a subset of CLL families. These findings are consistent with the evidence of linkage of familial CLL to chromosomes 7q31.32-q33 and 16q12.2-q23.1 that we previously observed (supplemental Figure 5).43
Germ line disruptive variants within shelterin genes have recently been implicated in predisposition to familial melanoma,39,40 cardiac angiosarcoma,44 glioma,45 and colorectal cancer,46 whereas somatic mutations of POT1 are detectable in 3.5% of all CLL and 9% of encoding immunoglobulin heavy chain variable-unmutated CLL,37 and were also identified in 10% of patients with cutaneous T-cell lymphoma.47
POT1-mutated CLL cells have numerous telomeric and chromosomal abnormalities, suggesting that POT1 mutation facilitates the acquisition of these malignant features.37 Our observation of germ line mutations in POT1 being associated with familial CLL would concur with this assumption. Our findings also support the proposal that POT1 mutation is an early event in CLL development.41
In a CLL GWAS, we previously reported an association between the common allele of the POT1 3′ untranslated region variant rs17246404 (risk allele frequency = 0.75) and increased CLL risk, with a small per allele effect size (odds ratio = 1.22).12 The recurrent POT1 coding variant, p.Gln376Arg, identified in the current study is not however in linkage disequilibrium with SNP rs17246404 (r2 = 0.00). Therefore, although further studies are required to determine exactly how rs17246404 influences CLL risk, it is plausible that the functional basis of the association is through differential gene expression.
Shelterin is a telomere-specific protein complex composed of 6 family members, encoded by POT1, ACD, TERF2IP, and TERF1, TERF2, and TINF2, that protects the ends of chromosomes. Together, the components of the shelterin complex are necessary for all telomere functions, including the protection of telomeres from degradation, aberrant recombination, and incorrect processing by DNA-repair machinery, as well as facilitating chromosome capping to mediate telomerase activity.48
POT1 directly contacts telomeric DNA overhangs49 and also binds to ACD,50 which connects POT1 to the other shelterin components via its bridge with TINF2.51 The POT1:ACD interaction enhances the affinity of POT1 for telomeric DNA.50,52 The ACD splice site mutation c.752-2A>C will disrupt the POT1 binding domain and abolish the TINF2 binding domain, so could therefore be predicted to result in an unformed shelterin complex.
In silico predictions suggest that the germ line p.Tyr36Cys mutation identified in pedigree 162 is likely to disrupt the interaction between POT1 and the single-stranded telomeric DNA overhang. The POT1 frameshift and splice site mutation are likely to result in truncated protein products, impairing their interaction with ACD. The p.Gln376Arg variant, though not predicted in silico to impact protein stability, alters an evolutionarily constrained residue thus implying functional importance.
Previous experiments have shown that when the ACD/POT1 subunit is inhibited, the telomerase complex increases telomere length.49,51 We observed no significant differences between the telomere lengths of CLL cases with a POT1 mutation and those who did not harbor a shelterin gene mutation. This observation is comparable to that in tumor cells derived directly from CLL cases with a somatic POT1 mutation vs matched cases with no POT1 mutation,37 and may reflect the numerous unmeasured variables that can influence telomere length in human populations. We also acknowledge that our telomere length measurements are based on blood-derived DNA and therefore could be subject to the effects of uncharacterized somatic mutations. In this regard, we note that the proband of pedigree 5047 harbored a somatic splice site mutation in ATR, a gene also known to play a key role in telomere maintenance.53 Furthermore, although GWAS have identified SNPs at loci including other telomere maintenance genes that are associated with telomere length, there has been no such association reported for a POT1 SNP.12,54,55
TERF2IP associates with the shelterin complex via its C-terminus to a central region of TERF2, forming a stable 1:1 complex. TERF2IP, as part of the shelterin complex, is vital for the repression of homology-directed repair of double-strand chromosomal break at the telomere. Although the novel missense variant p.Arg133Gln is predicted to be pathogenic, markedly reducing the stability of the protein, p.Ala104Pro is less well conserved and is thus more likely to be tolerated.
Germ line disruptive mutations in POT1 have previously been associated with susceptibility to melanoma in 9 families39,40 and glioma in 3 families.45 Furthermore, recent studies have identified POT1 p.Arg117Cys in 4 Li-Fraumeni–like syndrome families,44 and ACD and TERF2IP mutations in 8 melanoma families.56 None of the mutation carriers in the melanoma families featured cases of glioma or CLL. Similarly, the glioma families did not feature cases of melanoma or CLL and the only case of melanoma was seen in 1 of the Li-Fraumeni–like syndrome families. Collectively, these data and the fact that none of our families segregated glioma or melanoma, suggest that the penetrance associated with rare shelterin complex mutations is modest. Such an assertion is supported by our observation that the predicted deleterious p.Arg133Gln TERF2IP variant was identified in only 2 out of 3 CLL cases sequenced in family 4014. Additionally, in our case-control analysis, the POT1 p.Gln376Arg mutation was shown to confer a modest 3.6-fold increase in risk of CLL. Furthermore, the absence of significant LOH in the tumors of carriers, when examined,44 suggests that mutations in the shelterin complex genes do not function as high penetrance tumor suppressors but rather as moderate penetrance alleles.
Early age of onset in cancer can be indicative of inherited predisposition and it is noteworthy that in this study, mutation carriers were diagnosed with CLL much younger than the population average (59 years vs 71 years). Because 7 of the 66 CLL families were carriers of shelterin mutations, this translates to 11% of familial CLL being ascribed to mutations in this class of genes (95% CI, 4-21). However, we acknowledge that our analyses were based only on families ascertained in the United Kingdom, and therefore the impact of such mutations on familial CLL could vary depending on ethnicity. Moreover, it remains to be established, through additional studies, whether other CLL families are the consequence of polygenic susceptibility or as yet unidentified higher impact disease-causing mutations.
In conclusion, the POT1, ACD, and TERF2IP loss-of-function mutations we report here suggest that multiple components of the shelterin complex play a role in CLL predisposition. Moreover, they extend the spectrum of cancer associated with inherited mutations in these genes. It is however, likely that shelterin complex gene mutations confer cancer risks analogous to those associated with ATM heterozygosity57 or CHEK258 for breast cancer. Nevertheless, because the dysregulation of telomere protection has been identified as a target for potential therapeutic intervention in CLL, it may be possible that early identification of mutation carriers will facilitate improvements in future disease management.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful to all investigators, and the patients and individuals for their participation. This study made use of genotyping data on the 1958 BC; a full list of the investigators who contributed to the generation of these data is available at http://www.wtccc.org.uk/.
Principal funding for the study was provided by Bloodwise (LRF05001, LRF06002, and LRF13044). The authors acknowledge support from Cancer Research UK (C1298/A8362 supported by the Bobby Moore Fund), the Arbib Fund, and the Leicester Experimental Cancer Medicine Centre (C325/A15575 Cancer Research UK/UK Department of Health). B.K. received a doctoral studentship from the ICR, supported by the Sir John Fisher Foundation.
Authorship
Contribution: H.E.S. and R.S.H. drafted the manuscript; H.E.S. performed project management, sequencing, and bioinformatic analysis; B.K., D. Chubb, P.J.L., and K.L. performed bioinformatic analysis; P.B. performed sample preparation; S.J. performed sample database management; C.D., M.J.S.D., G.A.F., and D. Catovsky performed sample recruitment; and R.S.H. obtained financial support.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Richard S. Houlston, Division of Genetics and Epidemiology, The Institute of Cancer Research, 15 Cotswold Rd, Sutton, Surrey SM2 5NG, United Kingdom; e-mail: richard.houlston@icr.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal