Abstract
Follicular lymphoma (FL) is a generally incurable B-cell malignancy which has the potential to transform into highly aggressive lymphomas. Genomic studies indicate it is often a small subpopulation rather than the dominant population in the FL that gives rise to the more aggressive subtype.To resolve the underlying transcriptional networks of follicular B-cell lymphomas at single molecule and cell resolution, we leveraged droplet-based barcoding technology for highly parallel single cell RNA-Seq. We analyzed the transcriptomes from tens of thousands of cells derived from five primary FL tumors (average > 5,000 cells/sample). Simultaneously, we conducted multi-dimensional flow cell sorting to validate our characterizing of cellular lineages and critical expressed proteins. For each tumor, we identified multiple cellular subpopulations, matching known hematopoietic lineages. Despite some common features, such as MYC and BCLoverexpression, distinct transcriptional patterns and regulatory programs were evident among the different tumors. Within each tumor, subpopulations of malignant cell transcripts were compared to matched normal B-cells (Figure 1).
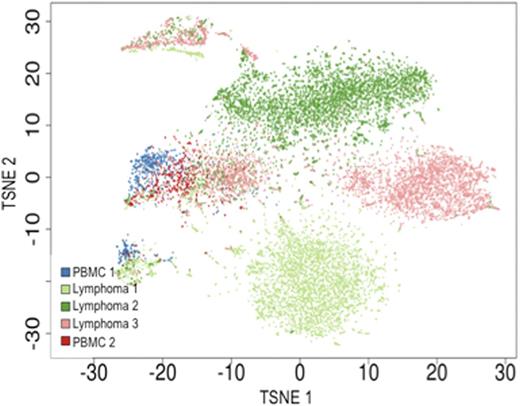
Transcriptome analysis of >13,500 B-cells across 5 samples reveals sample-specific and joint clusters of B-cells. Each dot represents the t-SNE projection of a single cell's transcriptome profile. Single cells are colored according to sample origin (control PBMC samples 1 & 2; follicular lymphoma samples 1, 2 & 3). Normal B-cells within follicular lymphoma samples cluster together with PBMC derived B-cells. Malignant B-cells cluster according to sample origin, except for the cycling tumor cells (upper left cell cluster).
Malignant B-cells were characterized by expression of restricted immunoglobulin light chain type (either kappa or lambda), BCL2 expression, and CD20 expression. We show evidence for the coexistence of small, malignant subpopulations alongside the dominant FL population in the majority of tumors. These smaller subpopulations harbored several transcriptional changes that were absent from the dominant population, including upregulation of MHC class II expression, downregulation of β-2-microglobulin, and exclusive expression of chemokines (CCL22 and CCL17), that would have resulted in alteration in antigen presentation and of the T cell immune system. In addition, we characterize the transcriptomes of the infiltrating immune cell populations that are characteristic of this disease. Our findings provide an unprecedented resolution of distinct immune lineages as seen by transcriptionally characterized cellular diversity.
Levy:Kite Pharma: Consultancy; Five Prime Therapeutics: Consultancy; Innate Pharma: Consultancy; Beigene: Consultancy; Corvus: Consultancy; Dynavax: Research Funding; Pharmacyclics: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal