Abstract
Background: While chronic myeloid leukaemia (CML) originates from a single genetic aberration (BCR-ABL1) remarkably heterogeneity characterises treatment response and outcome. Most CML patients respond well to tyrosine kinase inhibitors (TKI), particularly 2nd generation (2G) TKI but a significant minority shows resistance and a proportion experience progression. At diagnosis there are currently no biomarkers for patients at higher risk of progression who could be treated with more effective treatment or be selected for BMT at an early stage of therapy. Such biomarkers may also provide useful prognostic information in addition to the most valid biomarker to date, the BCR-ABL1 IS ratio during the first 3-6-12 months of TKI therapy.
Aims: The aim of our study is to analyse a panel of mutations in epigenetic modifiers in pre-treatment CML-CP using Ion Torrent next-generation sequencing (IONT-NGS) to assess the prognostic value of potential mutations as novel biomarkers of response to 1st and 2G TKI and risk of progression to advanced phase disease.
Methods: 100 samples from untreated patients with newly diagnosed CML-CP were included in the study, 62 from patients treated frontline with imatinib (IM) and 38 with a 2G-TKI (31 dasatinib, 4 nilotinib, 3 bosutinib). The patients were classified as TKI responders (R) (34 IM, 22 2G-TKI) or non-responders (NR) (28 IM, 16 2G-TKI) based on BCR-ABL1 IS ratio at 3 months. DNA was extracted from CD34+ cells isolated from diagnostic samples, while DNA from T cells was used as constitutional non-leukemic control to exclude confounding germline mutations. Samples from healthy donors (n=14) and CML blast crisis (BC) (n=5) patients were used as negative and positive controls, respectively. We used a custom panel covering the coding region of 71 epigenetic enzymes. After sequencing data processing was performed excluding variants of low quality, common in the general population with minor allele frequency (maf) >1% or present in the healthy controls, we analysed the genomic data and integrated them with annotated clinical data.
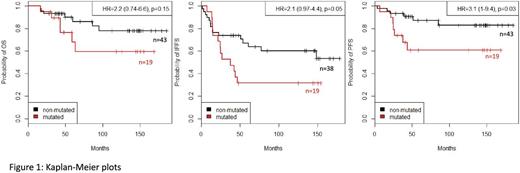
Results: After using a variant reduction pipeline, 164 non-synonymous variants that affected protein function were identified: 52 somatic and 112 germline. The somatic mutations (including missense, nonsense, frameshift insertions and splice site variants) were confined to 30 genes, with ASXL1, IKZF1, CREBBP beingthe most frequently mutated (n=9, 7 and 4 respectively). The mutations were detected in 34/100 (34%) CML-CP patients (19/62 IM and 15/38 2G-TKI), in higher proportion in NR (19/44, 43%) compared to R (15/56, 27%; p=0.027). We next correlated the presence of mutations with overall survival (OS), TKI failure free survival (TFFS) and progression free survival (PFS). IM patients carrying somatic mutations demonstrated a poorer response to IM [HR=2.1 (1-4.4 95% CI), p=0.05] and were more likely to progress to advanced phase [HR=3.1 (1-9.4 95% CI), p=0.03] (Figure 1). Nonsense mutations in particular (in ASXL1, IKZF1, DNMT3A, EP300) were found in 4 IM NR vs 1 R and their presence led to poor OS [HR= 6.1 (1.6-23 95% CI), p=0.002] and PFS [HR= 5.4 (1.4-21 95% CI), p=0.006]. As these were observed in 5 patients, further testing is required to corroborate this initial observation. Multivariate analysis revealed that both increased Sokal score and occurrence of somatic mutations negatively influenced outcome: somatic mutations detected in 6/24 low and in 8/13 high Sokal IM patients were associated with worse OS and PFS compared to unmutated patients with the same Sokal score. Among 38 patients treated with 2G-TKI, neither somatic mutations (including nonsense variants) nor combination of somatic mutations with Sokal score had any influence on OS, TFFS, PFS, neither did the presence of germline mutations in either IM or 2G-TKI patients.
Summary/Conclusion: Somatic mutations identified using IONT-NGS on 71 epigenetic modifiers potentially predict 1st generation (IM) patient poor survival, drug failure and progression to advance phase disease. However, the more effective therapeutic effect of 2G-TKI seems to overcome the poor prognostic influence of such mutations though further validation on larger cohort of patients may be required to validate preliminary data. Our results suggest that occurrence of somatic mutations at diagnosis have the potential to identify patients who would benefit from upfront treatment with 2G-TKI.
Apperley:Incyte: Speakers Bureau; Novartis: Honoraria, Speakers Bureau; Ariad: Honoraria, Speakers Bureau; Pfizer: Honoraria, Speakers Bureau; Bristol Myers Squibb: Honoraria, Speakers Bureau.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal