Abstract
Background: Hereditary spherocytosis (HS) is the most common cause of hereditary hemolytic anemia. Current tests used to diagnose HS focus on the detection of hemolysis or indirectly assess protein defects. Direct methods to detect protein defects are complicated and difficult to implement. Recent next-generation sequencing (NGS) methods enable large-scale gene mutation analyses to be used for such diagnoses. In this study, we investigated the patterns of genetic variation associated with HS among the patients diagnosed with HS clinically. Specifically, we analyzed mutations in red blood cell membrane protein-encoding genes (17 genes) in context with 5 genes for the differential diagnosis (thalassemia, congenital dyserythropoietic anemia, paroxysmal nocturnal hemoglobinuria) in Korean HS.
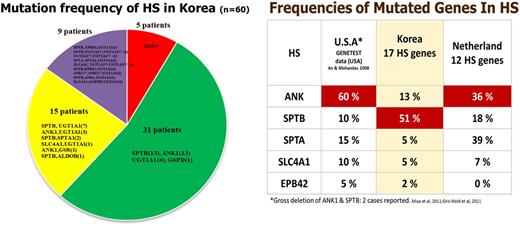
Methods: In total, 60 patients diagnosed with HS were enrolled in this study. Targeted sequencing of 43 genes (17 membrane protein-encoding genes, 20 enzyme-encoding genes, and 6 additional candidate genes) was performed using the Illumina HiSeq platform and variants were called according to a data-processing pipeline.
Results: Of the 60 patients, 50 (83%) had one or more significant variants in a membrane protein encoding genes. A total of 54 significant variants (8 previously reported and 46 novel) were detected in 6 membrane protein-encoding genes; SPTB, ANK1, SPTA1, SLC4A1, EPB41, and EPB42. The most variants (28/60 patients) were detected in SPTB. Interestingly, concurrent mutations of genes encoding enzymes (ALDOB, GAPDH, and GSR) were detected along with mutations of membrane encoding genes. One patient diagnosed with HS harbored mutation of G6PD without mutation of HS related genes. Additionally, UGT1A1 mutations were present in 5 patients. Positive rate of osmotic fragility test was 86% among patients with HS related gene mutations.
Conclusion: These results clarify the molecular genetic analysis is required for the accurate diagnosis of HS. About 17% of patients who were clinically diagnosed as HS revealed discrepancy with molecular diagnosis.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal