Abstract
Introduction
Chronic myeloid leukemia (CML) patients who do not achieve landmark responses are considered imatinib (IM)-resistant. IM-resistance can be due to BCR-ABL kinase domain (KD) mutations, although many IM-resistant patients do not harbor KD mutations. The pathogenesis of this phenomenon is unclear. Aberrations in BCR-ABL independent downstream signaling pathways, namely PI3K/AKT, p53, NF-kB and Fanconi anaemia (FA)/BRCA, have been implicated. MicroRNAs (miRNAs) are short non-coding RNAs that control gene expression and are notoriously promiscuous, with one miRNA regulating many mRNAs. In recognition of this, we sought to identify dysregulated miRNAs associated with the above pathways in patients with IM-resistant CML.
Methods
Eight patients with chronic phase CML who demonstrated primary resistance to IM and tested negative for KD mutations via Sanger sequencing were enrolled. Two healthy volunteers constituted the control group. Peripheral blood samples were taken and miRNA extracted from the white cells. MiRNA profiling was performed using miRNA sequencing (miRNA-seq) of the human microRNAome. MiRNA expression analyses (miEA) were performed on 4 sets of miRNAs associated with 4 different signaling pathways derived from publicly available databases: 237 miRNAs for PI3K/AKT, 223 miRNAs for p53, 221 miRNAs for NF-kB, and 126 miRNAs for FA/BRCA. MiRNA expression levels of the patients were compared with normal controls to obtain the differential expression of the miRNAs. MiRNAs that occur in all 4 pathways were further investigated in silico for their putative gene targets (Tarbase v7.0). Gene targets which were commonly linked with 3 different miRNAs were identified.
Results
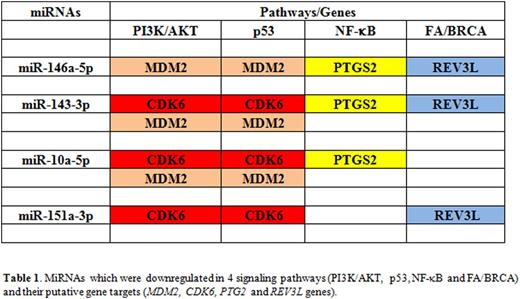
MiRNA-seq yielded a total of 790 miRNAs. MiEA showed downregulation of 5 miRNAs in all 4 signaling pathways above: miR-146a-5p, miR-99b-5p, miR-143-3p and miR-10a-5p, miR-151a-3p. MiR-146a-5p and miR-99b-5p were the first and second most downregulated miRNAs in all 4 pathways. Putative gene targets that were commonly modulated by 3 miRNAs were: CDK6 (miR-143-3p, miR-10a-5p, miR-151a-3p), MDM2 (miR-146a-5p, miR-143-3p, miR-10a-5p), PTGS2 (miR-146a-5p, miR-143-3p, miR-10a-5p) and REV3L (miR-146a-5p, miR-143-3p, miR-151a-3p), as shown in Table 1.
Conclusions
The PI3K/AKT pathway is activated in response to IM-exposure to IM-naïve cells, implying a role in IM resistance. As cell cycle modulators, both p53 and NF-kB pathways are regulated within the PI3K/AKT pathway via AKT. FANCD2 is a central FA/BRCA pathway regulator and its transcription is maintained by AKT/mTOR via enhancement of NF-kB activity. The complexity of the interactions above has impeded efforts in identifying a unifying target in counteracting IM resistance. Thus, it is of significance that our results showed 5 differentially expressed miRNAs that consistently featured in the above signaling pathways, 4 of which remarkably influence PI3K/AKT and p53.
In our study, PI3K/AKT and p53 pathways are inextricably linked with the same 4 miRNAs (miR-146a-5p, miR-143-3p, miR-10a-5p and miR-151a-3p) targeting the same 2 genes (MDM2 and CDK6) in these 2 pathways. MDM2 is an ubiquitin ligase that regulates the stability of p53a. AKT facilitates the functions of MDM2 to promote p53 ubiquitination. Conversely, inhibition of MDM2 leads to activation of p53 in response to cellular stresses. CDK6 is a cell cycle regulator and its overexpression influences the accumulation of pro-apoptotic p53 proteins, postulating a role in DNA damage response. Both PTGS2 and REV3L genes were commonly targeted by 3 miRNAs, within the NF-kB and FA/BRCA pathways respectively. PTGS2 overexpression has been shown in gastrointestinal adenocarcinomas; it induces carcinoma cell migration and invasion via activation of PI3K/AKT signaling. REV3L is the catalytic subunit of DNA polymerase ζ and plays a central role in DNA damage tolerance and chemoresistance towards DNA damaging agents. Notably, the NF-kB and FA/BRCA pathways were juxtaposed to the PI3K/AKT/p53 system via shared miRNA regulators.
This provokes interesting questions regarding IM-resistance: are anti-apoptotic signaling and DNA damage response mechanisms driven simultaneously by these miRNAs? Can these miRNAs be potential biomarkers and therapeutic targets in IM-resistant CML? More studies are needed to further validate our findings.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal