Abstract
Anucleated platelets have a rich repertoire of RNAs, many of which have provided insights into megakaryocyte-platelet biology and have been proposed as disease biomarkers. Although it is generally believed that the platelet transcriptome is a reflection of the megakaryocyte (MK) RNA content and is transported into platelets during pro-platelet production, this is a poorly studied concept. Healthy human bone marrow (BM) is difficult to access and because MKs represent <0.05% of nucleated cells in BM, making it technically challenging to obtain a pure population. Cultured MKs from CD34-positive umbilical vein cord blood have advantages of being human and initially primary cells, but it is unknown how well the transcriptomes of differentiating MKs in culture correlate with primary BM MKs. Microdissection of BM aspirates by laser capture scanning microscopy can overcome purity limitations by isolating virtually 100% MKs. The goals of this study were to assess the transcriptome correlations between laser-captured primary human MKs (LC-MKs) and both (1) platelets from the same individuals who donated the BM and (2) cord blood derived MKs (CB-MKs).
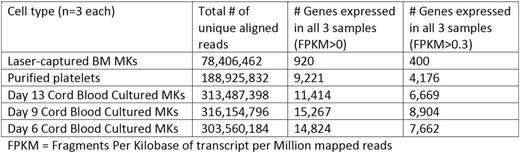
BM aspirates and peripheral blood were collected from three healthy donors. Mature BM MKs were identified by a Board certified Hematologist using standard morphologic criteria, and approximately 200 MKs were microdissected by laser capture scanning microscopy from each aspirate. Peripheral blood platelets were isolated from the same donors followed by leukocyte and erythrocyte depletion. Separately, CD34-positive cells were isolated from umbilical vein cord blood, cultured under standard MK-producing conditions with thrombopoietin and stem cell factor, and CD61-positive CB-MKs were isolated at day 6, 9 and 13 in three independent experiments. Total RNA was extracted from LC-MKs, platelets and CB-MKs and subjected to next-generation RNA-sequencing (total of 15 samples: 5 cell types from 3 independent experiments). The Table summarizes the RNA-seq read numbers generated and the number of protein-coding genes determined for each sample. Notably, the LC-MK RNA yielded fewer reads because the input (estimated in the low picogram range) was less than the other samples. Several known platelet transcripts, including PPBP, PF4, B2M, TMSB4X and OST4 were abundant in our LC-MKs RNA-sequencing analysis. All samples exhibited high expression (1,721-74,923 FPKM) of mitochondrial transcripts.
To assess RNA correlations across cell types, we only considered transcripts expressed in both comparison groups. mRNAs from LC-MKs were statistically significantly correlated with mRNAs from both platelets and day 13 CD41a-CD42a-positive CB-MKs. These correlations were highest for the most abundantly expressed (FPKM>50) transcripts (Spearman rank correlation Rho = 0.71 for LC-MKs vs. platelets, and Rho = 0.67 for LC-MKs vs. day 13 CB-MKs). However, even the lower expressed transcripts (using the standard threshold of FPKM>0.3) showed moderate correlation (Rho = 0.37-0.42, p value <E-13). Regardless of transcript abundance, the platelet transcriptome displayed higher correlations with LC-MKs than did CB-MKs. Of the 400 mRNAs common to all LC-MKs samples, 23 were not identified in any of the platelet samples and 9 were not identified in any of the d13 CB-MKs. A secondary goal was to compare transcriptome profiles of CB-MKs during differentiation. Not surprisingly, mRNAs from cultured CB-MKs showed higher correlation when their time in culture was shorter than longer (Rho = 0.87 for d13 vs. d9; 0.81 for d9 vs. d6; and 0.64 for d13 vs. d6). During CB-MK differentiation, 825 transcripts were up-regulated whereas 2725 transcripts were down-regulated.
In summary, we provide the first report of transcript profiling from normal, primary human MKs. Peripheral blood platelet RNA transcript levels were representative of MK transcripts from the same subjects, especially if abundantly expressed; cultured MK mRNAs correlated strongly with LC-MKs, while platelet RNAs from the same donor showed an even stronger correlation. mRNA expression differences between platelets and BM MKs could provide insights into thrombopoiesis and may be due to (1) regulated transcript partitioning from MK to platelets, (2) differential RNA endocytosis or exocytosis or (3) RNA degradation.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal