Key Points
FLs harbor more recurrent mutations in the BCR signaling pathway, SWI/SNF complex, and histone genes than previously known.
Novel recurrent mutations affecting BTK, SYK, and HVCN1 may have therapeutic and prognostic implications for FL.
Abstract
Follicular lymphoma (FL) is the most common form of indolent non-Hodgkin lymphoma, yet it remains only partially characterized at the genomic level. To improve our understanding of the genetic underpinnings of this incurable and clinically heterogeneous disease, whole-exome sequencing was performed on tumor/normal pairs from a discovery cohort of 24 patients with FL. Using these data and mutations identified in other B-cell malignancies, 1716 genes were sequenced in 113 FL tumor samples from 105 primarily treatment-naive individuals. We identified 39 genes that were mutated significantly above background mutation rates. CREBBP mutations were associated with inferior PFS. In contrast, mutations in previously unreported HVCN1, a voltage-gated proton channel-encoding gene and B-cell receptor signaling modulator, were associated with improved PFS. In total, 47 (44.8%) patients harbor mutations in the interconnected B-cell receptor (BCR) and CXCR4 signaling pathways. Histone gene mutations were more frequent than previously reported (identified in 43.8% of patients) and often co-occurred (17.1% of patients). A novel, recurrent hotspot was identified at a posttranslationally modified residue in the histone H2B family. This study expands the number of mutated genes described in several known signaling pathways and complexes involved in lymphoma pathogenesis (BCR, Notch, SWitch/sucrose nonfermentable (SWI/SNF), vacuolar ATPases) and identified novel recurrent mutations (EGR1/2, POU2AF1, BTK, ZNF608, HVCN1) that require further investigation in the context of FL biology, prognosis, and treatment.
Introduction
Follicular lymphoma (FL) is the most common indolent non-Hodgkin lymphoma (iNHL), and is characterized by a variable clinical course.1,2 Although patients with FL typically respond to standard therapies, this disease remains largely incurable even when considering therapeutic advances.3 The varied clinical course of FL is reflected in an approximately 30% lifetime risk of transforming from an iNHL to a more aggressive lymphoma, most commonly diffuse large B-cell lymphoma, which is associated with poor prognosis.4 In contrast, some patients may be safely observed for decades, not requiring treatment years after initial diagnosis.2 The pathobiology of FL is complex and involves cell-intrinsic genetic changes [eg, the hallmark t(14;18) translocation resulting in BCL2 overexpression], as well as alterations within the FL microenvironment.5,6 Although recent studies using next-generation sequencing have illuminated genetic perturbations that occur in FL, especially those linked to transformation,7-10 our understanding of the genomic landscape of FL remains incomplete.
A number of studies have focused on discovering recurrent genomic alterations in B-cell NHL; however, early studies included only limited analysis of FL samples.11 Despite a small sample set, several recurrent somatic mutations were identified involving the EZH212 and highly recurrent KMT2D (MLL2) genes.13 More recently, several studies have performed exome-based discovery sequencing on small sets of patients with FL, with subsequent extension sequencing of highly recurrent mutations in larger validation cohorts.14-17 These approaches have confirmed the impact of mutations previously identified and revealed a number of recurrently mutated genes and pathways in FL, including histone modifiers, histone H1 genes, B-cell receptor signaling genes, STAT6, POU2F2, and others. Using a panel of 74 genes, Pastore et al18 identified 7 recurrently mutated genes that added to the predictive ability of the standard clinical FL international prognostic index (FLIPI), resulting in the new m7-FLIPI. These important findings, based on small sample sets and focused gene panels, have laid the groundwork for our understanding of the pathobiology of FL. To build on this work, broader strategies targeting more genes in larger cohorts are needed to identify less common, significantly mutated genes (SMGs) and other genes within known pathways that contribute to the heterogeneous nature of FL pathobiology.
To better define the genetic landscape of FL, identify genes with a statistically significant rate of mutation in FL, and improve our comprehension of pathways altered in FL, exome sequencing was performed on samples from a discovery cohort of 24 patients with FL and matched normal tissue. Leveraging the combination of this discovery cohort and existing sequencing studies of B-cell malignancies,12,13,19-26 we designed a custom capture panel to sequence 1716 genes in 113 samples from primarily treatment-naive patients with FL. This approach identified novel, recurrently mutated genes in known and putative lymphoma-associated cellular pathways.
Methods
Patient characteristics and sample acquisition
For the discovery cohort, all patients provided written informed consent for the use of their samples in sequencing as part of the Washington University School of Medicine (WUSM) Lymphoma Banking Program. Excisional biopsy tissue and nonmalignant (skin punch biopsies) samples were collected (2008-2013). Pathology review was performed on frozen lymph node samples to confirm the diagnosis and estimate tumor cell content. Frozen sections (tumor and skin) were cut and used for genomic DNA isolation. Flow sorting was performed on a Reflection instrument. For the extension cohort, FFPE blocks from an excisional biopsy were identified from patients evaluated at WUSM/Siteman Cancer Center (2001-2013), reviewed to confirm the pathology diagnosis, and marked for block punches in confirmed tumor-containing areas. All samples were collected within protocols approved by the WUSM institutional review board (201108251, 201104048, 201110187).
Library preparation and sequencing
Genomic DNA was isolated using the QIAamp DNA Mini kit, with xylene or CitroSolv to remove paraffin from FFPE samples. Library preparation, capture hybridization, and sequencing using a HiSeq 2000 were performed as previously described.27 Library capture was performed using a NimbleGen SeqCap EZ Exome v2.0 or 7.05-Mb NimbleGen Custom Liquid Capture reagent (supplemental Table 2, available on the Blood Web site). Sequencing data have been deposited (dbGaP accession: phs001229.v1).
Sequence alignment and variant calling
Sequence alignment was performed using the Genome Modeling System.28 Briefly, paired-end reads were aligned to human reference sequence GRCh37, using BWA29 (discovery) or BWA-MEM30 (extension), and de-duplicated using Picard (http://broadinstitute.github.io/picard/). Variants were identified using SAMtools,29 SomaticSniper,31 VarScan,32 MuTect,33 Strelka,34 Pindel,35 and GATK,36 and annotated using the GMS variant annotator (Ensembl v74). Variants were filtered to remove common variants37-39 and pipeline artifacts. See supplemental Methods for more extensive details. All analyses involving the extension cohort were restricted to nonsynonymous and splice site mutations.
Variant analysis
Significantly mutated genes (SMGs) were identified using MuSiC v0.4, with the false discovery rate (FDR) cutoff based on the convolution test method.40 Mutual exclusivity and associated mutations were determined by permutation analysis. The estimated P-value was determined using the number of permuted data sets with more total co-occurring mutations than the real data set over 10 000 permutations (supplemental Methods). Visuals were created using GenVisR.41
Histone gene analysis
Univariate clinical analysis
A log-rank test was used for survival analysis with patient groups stratified by FLIPI score or mutation status (SMGs with 5 coding/splice site or 3 truncating mutations). Clinical associations with SMGs were tested using a χ-squared or Fisher’s exact test (supplemental Methods). The Benjamini-Hochberg method was used for multiple testing corrections. Statistical analysis was performed using R (v3.2.1) with “survival,” “multtest,” and “stat” packages.
Results
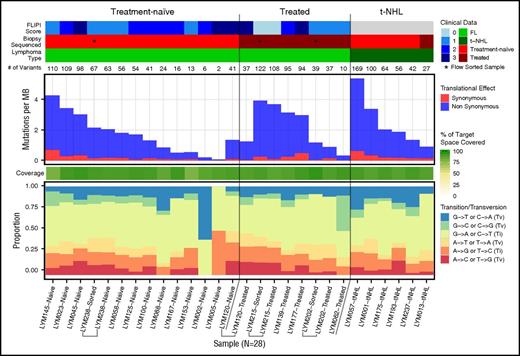
To identify novel recurrent mutations in FL, we performed exome sequencing of 28 fresh-frozen tumor samples with paired nonmalignant (skin) tissue from 24 patients. This included 13 patients with treatment-naive FL (1 with paired relapse and 1 with a sample derived from both bulk lymph node and after flow-sorting to purify to more than 98% light chain-restricted CD19+ lymphoma cells), 5 with relapsed disease (2 with flow-sorted cells), and 6 with transformed FL (Figure 1; supplemental Table 1). Flow sorting of lymphoma cells resulted in similar variant detection in 2 samples and improved variant detection in a third, compared with bulk lymph node tissue (supplemental Figure 1). The performance of both bulk frozen and FFPE samples was sufficient for analyses, although flow sorting may improve the accurate detection of low-frequency subclonal mutations in future studies. In the 1 patient with longitudinal samples (diagnosis and relapse) available, a founding clone with acquisition of a small number of mutations at relapse was evident (supplemental Figure 1). The median number of mutations per megabase was 1.755 (range, 0.066-5.372), with a median of 55 mutations (range, 2-169) per individual.
Mutation numbers and spectrum within the FL discovery sample set. Baseline genomic features of FL are shown for the exome sequenced discovery cohort. Clinical features (upper) are indicated for all 28 samples sequenced from 24 individuals. Immediately below the clinical features is a row indicating the total number of mutations per sample. Mutations per megabase sequenced (middle) is based on the total mutations within the targeted exome capture space successfully covered in each sample (80% breadth, 20× depth), with the percentage of the sequenced target region covered in each sample indicated immediately below in green. Finally, the rate of transitions and transversions in the mutations observed in each individual are shown (bottom). Bulk fresh-frozen samples were sequenced unless indicated (*) as a flow-sorted sample. Brackets group multiple samples from a single individual. MB, megabase; Ti, transition; Tv, transversion
Mutation numbers and spectrum within the FL discovery sample set. Baseline genomic features of FL are shown for the exome sequenced discovery cohort. Clinical features (upper) are indicated for all 28 samples sequenced from 24 individuals. Immediately below the clinical features is a row indicating the total number of mutations per sample. Mutations per megabase sequenced (middle) is based on the total mutations within the targeted exome capture space successfully covered in each sample (80% breadth, 20× depth), with the percentage of the sequenced target region covered in each sample indicated immediately below in green. Finally, the rate of transitions and transversions in the mutations observed in each individual are shown (bottom). Bulk fresh-frozen samples were sequenced unless indicated (*) as a flow-sorted sample. Brackets group multiple samples from a single individual. MB, megabase; Ti, transition; Tv, transversion
From the 2262 variants identified in this analysis, we selected 898 mutated genes to target, using a custom capture reagent. To these, we added 818 genes recurrently mutated (>1) in samples from 10 sequencing studies of other B-cell non-Hodgkin lymphomas (supplemental Tables 2 and 3) and the 3′UTR of BCL2. This WUSM lymphoma panel (WUSM-LP) was applied to the 28 discovery samples and an additional 83 FL samples from 81 individuals (71 treatment-naive, 7 posttreatment, 1 transformed, 2 treatment status unknown; supplemental Table 1; supplemental Figure 2). In total, 113 tumor samples (36 with paired normal samples) from 105 individuals were sequenced, using a targeted panel of 1716 genes, achieving greater than 20× coverage for more than 75% of the targeted region for all samples (Table 1; supplemental Figure 3; supplemental Table 4).
Clinical characteristics of patients used for genetic and clinical analysis
| Characteristic . | Genetic analysis value . | Clinical analysis value . |
|---|---|---|
| Total patient number | 105 | 82 |
| Female, % | 54.3 | 56.1 |
| Male, % | 45.7 | 43.9 |
| Age (median) | 58 | 58.5 |
| Age range | 22-87 | 28-87 |
| Stage, % | ||
| I | 19.0 | 15.9 |
| II | 5.7 | 7.3 |
| III | 27.6 | 28.1 |
| IV | 44.8 | 48.8 |
| NA: no information | 2.9 | — |
| FLIPI score, % | ||
| Low | 36.2 | 36.6 |
| Intermediate | 35.2 | 41.5 |
| High | 20.0 | 22.0 |
| NA: no information or tNHL | 8.6 | — |
| m7 FLIPI score, % | ||
| Low | 75.2 | 87.7 |
| High | 11.4 | 12.4 |
| NA: no information or tNHL | 13.3 | — |
| Lymphoma type, % | ||
| FL | 91.4 | — |
| Transformed lymphoma (tNHL) | 6.7 | — |
| NA: no information | 1.9 | — |
| Sequenced biopsy, % | ||
| Treatment-naive FL | 80.0 | 100.0 |
| Treated FL* | 11.4 | — |
| Transformed lymphoma (tNHL) | 6.7 | — |
| NA: no information | 1.9 | — |
| Treatment, % | ||
| Rituximab containing regimen | — | 50.0 |
| Other treatment† | — | 22.0 |
| Observation | — | 28.1 |
| Best response to treatment, %‡ | ||
| Complete remission | — | 66.1 |
| Partial remission | — | 28.8 |
| Stable disease | — | 1.7 |
| Progressive disease | — | 3.4 |
| Characteristic . | Genetic analysis value . | Clinical analysis value . |
|---|---|---|
| Total patient number | 105 | 82 |
| Female, % | 54.3 | 56.1 |
| Male, % | 45.7 | 43.9 |
| Age (median) | 58 | 58.5 |
| Age range | 22-87 | 28-87 |
| Stage, % | ||
| I | 19.0 | 15.9 |
| II | 5.7 | 7.3 |
| III | 27.6 | 28.1 |
| IV | 44.8 | 48.8 |
| NA: no information | 2.9 | — |
| FLIPI score, % | ||
| Low | 36.2 | 36.6 |
| Intermediate | 35.2 | 41.5 |
| High | 20.0 | 22.0 |
| NA: no information or tNHL | 8.6 | — |
| m7 FLIPI score, % | ||
| Low | 75.2 | 87.7 |
| High | 11.4 | 12.4 |
| NA: no information or tNHL | 13.3 | — |
| Lymphoma type, % | ||
| FL | 91.4 | — |
| Transformed lymphoma (tNHL) | 6.7 | — |
| NA: no information | 1.9 | — |
| Sequenced biopsy, % | ||
| Treatment-naive FL | 80.0 | 100.0 |
| Treated FL* | 11.4 | — |
| Transformed lymphoma (tNHL) | 6.7 | — |
| NA: no information | 1.9 | — |
| Treatment, % | ||
| Rituximab containing regimen | — | 50.0 |
| Other treatment† | — | 22.0 |
| Observation | — | 28.1 |
| Best response to treatment, %‡ | ||
| Complete remission | — | 66.1 |
| Partial remission | — | 28.8 |
| Stable disease | — | 1.7 |
| Progressive disease | — | 3.4 |
One patient with both treatment-naive and treated biopsies was only counted as treatment-naive.
Other treatment includes: CHOP (cyclophosphamide, hydroxydaunomycin, oncovin, prednisolone), bendamustine-ofatumumab + ofatumumab, XRT (radiation therapy), bendamustine + ofatumumab, cyclophosphamide, CVP (cyclophosphamide, vincristine, prednisolone), CVP + genitope protocol vaccine, and CHOP + XRT.
This excludes patients who were observed.
Somatic mutation analysis and confirmation of recurrent gene mutations
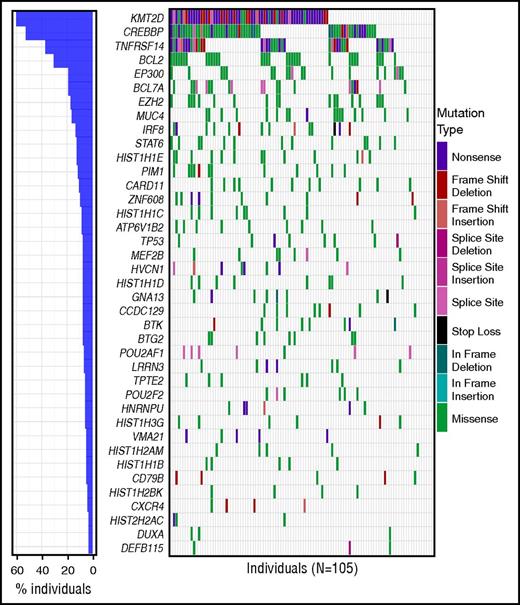
After filtering to remove common variants and pipeline artifacts, the number of nonsynonymous coding variants (including splice donor/acceptor sites) per sample ranged from 1 to 116 (median, 40). Of the 1468 affected genes, 889 were mutated in more than 1 patient (supplemental Table 5). We identified 39 genes that were significantly mutated above background mutation rates (herein referred to as SMGs) within the custom capture space (FDR < 0.05) (Figure 2; supplemental Table 6). This included many genes with established relevance to FL, including 5 previously described chromatin/epigenetic modifiers (KMT2D/MLL2 [60.0% mutation rate in this cohort], CREBBP [52.4%], EP300 [19.0%], EZH2 [17.0%], MEF2B [7.6%])12,13,44 and other established recurrently mutated genes (IRF8 [13.3%], STAT6 [12.4%], BTG2 [8.5%], and PIM1 [11.4%]; Figure 2).13,15,16 Supplemental Figures 4 and 5 illustrate the distribution of these mutations across samples stratified by clinical and sequencing parameters. Comparing treatment-naive samples (N = 84) with either t-NHL (N = 7) or posttreatment samples (N = 13) showed overrepresentation of TP53 (P = .005; q = 0.891) and EP300 (P = .032; q = 0.966) mutations in the t-NHL samples and HIST1H3G mutations (P = .030; q = 0.966) in posttreatment samples (supplemental Table 7). However, these results are merely suggestive, as the majority of samples were treatment-naive, limiting the power of this analysis, and were not significant after multiple hypothesis testing. Many individuals (N = 25) harbored more than 1 KMT2D mutation, with 89 mutations observed in 63 individuals (supplemental Figure 6). Mutations in STAT6 primarily occurred at the D419 position, which has been shown to result in activation, with a frequency consistent with previous reports.8,16 However, we did not observe complete overlap with CREBBP mutations, as previously reported, as 4/13 patients harbored STAT6 mutations alone (supplemental Figure 7).
SMGs in FL. The frequency and type of mutations affecting 39 genes identified as significantly mutated in our cohort using MuSiC analysis (FDR < 0.05, convolution test method) are displayed in each row. Columns represent each patient in the cohort and are ordered by the presence of mutations in the most to least frequently mutated gene. The bar graph on the left corresponds to the frequency of mutations for that gene in the entire cohort. For genes with multiple mutations in a single patient, only 1 mutation type is shown with priority order indicated in the legend from the highest priority at the top to lowest at the bottom. For individuals with multiple samples, the union of mutations in all samples for that individual was used. The mutation waterfall plot was created using the “GenVisR” package in R.41
SMGs in FL. The frequency and type of mutations affecting 39 genes identified as significantly mutated in our cohort using MuSiC analysis (FDR < 0.05, convolution test method) are displayed in each row. Columns represent each patient in the cohort and are ordered by the presence of mutations in the most to least frequently mutated gene. The bar graph on the left corresponds to the frequency of mutations for that gene in the entire cohort. For genes with multiple mutations in a single patient, only 1 mutation type is shown with priority order indicated in the legend from the highest priority at the top to lowest at the bottom. For individuals with multiple samples, the union of mutations in all samples for that individual was used. The mutation waterfall plot was created using the “GenVisR” package in R.41
Highly recurrent histone gene mutations often co-occur in patients with FL
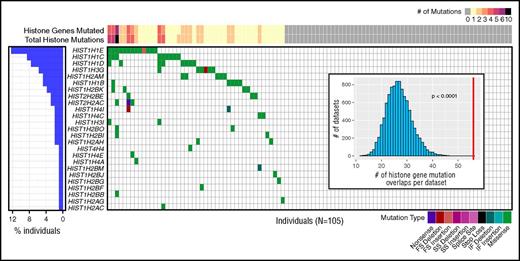
Recent studies have identified recurrent mutations in histone H1 genes in FL.8,15 In our sequencing data, 8 histone genes (HIST1H1B, HIST1H1C, HIST1H1D, HIST1H1E, HIST1H2AM, HIST1H2BK, HIST1H3G, and HIST1H2AC) were significantly mutated, and a total of 25 histone genes harbored at least 1 coding mutation across the cohort, affecting 46 (43.8%) patients. Eighteen patients (17.1%) had mutations in more than 1 histone gene. Evaluation of the mutual exclusivity or association of these mutations using permutation analysis showed that these mutations co-occurred within patients more often than expected by chance (estimated P-value < .0001; Figure 3; supplemental Figure 8). We verified these co-occurring mutations were not mapping artifacts caused by homology (supplemental Table 8; supplemental Methods).
Histone gene mutations co-occur within individual patients with FL. Coding and splice site mutations in genes encoding the core histones (H2A, H2B, H3, H4) or histone linker (H1) often co-occur within patients. Each row represents a mutated histone gene, and each column represents a patient in this cohort. Histone mutations per patient are displayed at the top, indicating the total number of genes mutated (also summarized for the cohort in the bar graph on the left) and total number of mutations observed (includes multiple mutations per gene). The distribution of mutations and mutation types are indicated by colored boxes in the grid. For genes with multiple mutations in a single patient, only 1 mutation type is shown, with priority order indicated in the legend from the highest priority at the left to lowest at the right. Visualization created using GenVisR.41 (Inset) Histogram depicts the distribution of expected total histone gene mutation co-occurrences from 10 000 randomly permutated datasets with respect to the observed total co-occurrence in this cohort indicated by a red line (estimated P value < .0001). Although some patients had more than 1 mutation per histone gene, as indicated at the top, genes were considered mutated or not mutated for co-occurrence analysis. See supplemental Table 11 for a complete list of mutations. FS, frame shift; IF, in frame; SS, splice site.
Histone gene mutations co-occur within individual patients with FL. Coding and splice site mutations in genes encoding the core histones (H2A, H2B, H3, H4) or histone linker (H1) often co-occur within patients. Each row represents a mutated histone gene, and each column represents a patient in this cohort. Histone mutations per patient are displayed at the top, indicating the total number of genes mutated (also summarized for the cohort in the bar graph on the left) and total number of mutations observed (includes multiple mutations per gene). The distribution of mutations and mutation types are indicated by colored boxes in the grid. For genes with multiple mutations in a single patient, only 1 mutation type is shown, with priority order indicated in the legend from the highest priority at the left to lowest at the right. Visualization created using GenVisR.41 (Inset) Histogram depicts the distribution of expected total histone gene mutation co-occurrences from 10 000 randomly permutated datasets with respect to the observed total co-occurrence in this cohort indicated by a red line (estimated P value < .0001). Although some patients had more than 1 mutation per histone gene, as indicated at the top, genes were considered mutated or not mutated for co-occurrence analysis. See supplemental Table 11 for a complete list of mutations. FS, frame shift; IF, in frame; SS, splice site.
To investigate possible mutation hotspots in histone family genes, we mapped these mutations to alignments of histone protein reference sequences (supplemental Table 9). These alignments uncovered several locations at which mutations occurred at the homologous amino acid residue in multiple histone family members (supplemental Figure 9). Most strikingly, in the core histone H2B family, we saw mutations altering position S37 in 4 H2B protein-encoding genes. This H2B S37 residue and adjacent Y38 (also mutated in this cohort) are phosphorylated in response to stress and during cell division, respectively.45,46 Y38 phosphorylation mediated by WEE1, a tyrosine kinase recurrently mutated in Hodgkin lymphoma,47 was previously shown to inhibit transcription of core histone genes.46 Finally, we tested whether multiple mutations in the same histone gene clustered in 3-dimensional (3D) space, and identified significant clusters in HIST1H1E and HIST1H2BK (supplemental Table 10; supplemental Figure 9). The residues that form a significant 3D cluster in HIST1H1E are located in the H5 globular domain,48 including the K81 residue, which corresponds to 1 of several sites involved in binding to nucleosomal linker DNA.8,49 The HIST1H2BK cluster including the S37 residue is located in the histone fold domain, which is responsible for the formation of the nucleosome and DNA contacts.50 Collectively, the higher frequency of histone mutations than previously reported, significant co-occurrence within patients, identification of hotspots across histone gene families, recurrent mutations in conserved H2B phosphorylation sites, and identification of nonrandom clusters of mutations within 3D space support the growing body of data that suggests histone gene mutations play an important role in FL.8,10,15
Recurrent mutations in the B-cell receptor and CXCR4 signaling pathways
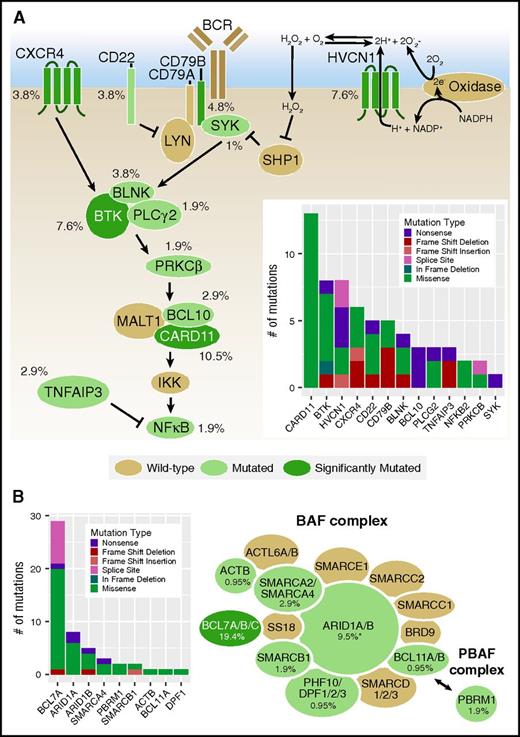
SMGs were identified in the interconnected B-cell receptor (BCR) and CXCR4 signaling pathways, including mutations in previously reported genes CD79B, CARD11, and CXCR413,18 (supplemental Tables 5 and 11), as well as novel mutations within HVCN1 (hydrogen voltage-gated channel 1) and BTK (Figure 4A). HVCN1 mutations were predominantly frameshift, nonsense, or splice-donor mutations, the significance of which is unclear, given its purported role for enhancing BCR signaling. The observed BTK mutations primarily occurred within the tyrosine kinase domain (supplemental Figure 10), including an L528W mutation that was previously associated with ibrutinib resistance in chronic lymphocytic leukemia (CLL)51 and an in-frame deletion that also alters this amino acid and the adjacent C527. No mutations affecting C481, the highly recurrent ibrutinib-resistance mutation observed in CLL, were noted; however, both C527 and L528W form a significant cluster with C481S in 3D space (supplemental Table 10).52 We also identified 2 BTK mutations, T117P and R562W, that are associated with X-linked agammaglobulinemia.53 It is not clear how these mutations will affect BTK in a somatic context, but they are likely to affect the function of BTK, and as a consequence, B-cell function as well. CARD11 mutations in 10 of 11 individuals occur within the coiled-coil domain, where missense mutations have been demonstrated to cause constitutive activation of NF-kappaB signaling in cell line assays.54 Four individuals in our cohort harbored 1 of the precise missense mutations shown to activate NF-kappaB in that study. Although not significantly mutated as individual genes, we also identified recurrent mutations in CD22, BLNK, BCL10, TNFAIP3, PRKCB, PLCG2, SYK, and NFKB2. Truncating BCL10 mutations observed in our cohort retain the CARD and MALT binding domains, similar to previously described truncating mutations, and thus may result in downstream NF-kappaB activation.55,56 Nonsense and frameshift mutations were observed in the NF-kappaB inhibitor, TNFAIP3, likely also leading to increased NF-kappaB signaling. In total, 47 patients (44.8%) harbored a mutation in the BCR pathway (supplemental Table 11).
Frequencies of mutations affecting the BCR/CXCR4 signaling pathways and SWI/SNF complex in patients with FL. (A) The interconnected BCR and CXCR4 signaling pathways are shown. Genes with nonsynonymous coding or splice site mutations are depicted in green, with SMGs in dark green and the mutation frequency observed in the entire cohort (N = 105) indicated. The total number and types of mutations observed are shown in the inset bar graph. (B) Recurrent mutations affecting both BAF (BRG1-associated factor) and PBAF (polybromo BRG1-associated factor) SWI/SNF complexes were observed in our cohort and annotated as in A. See supplemental Table 11 for a complete list of mutations. *Frequency includes 2 ARID1A variants rescued after ESP filtering.
Frequencies of mutations affecting the BCR/CXCR4 signaling pathways and SWI/SNF complex in patients with FL. (A) The interconnected BCR and CXCR4 signaling pathways are shown. Genes with nonsynonymous coding or splice site mutations are depicted in green, with SMGs in dark green and the mutation frequency observed in the entire cohort (N = 105) indicated. The total number and types of mutations observed are shown in the inset bar graph. (B) Recurrent mutations affecting both BAF (BRG1-associated factor) and PBAF (polybromo BRG1-associated factor) SWI/SNF complexes were observed in our cohort and annotated as in A. See supplemental Table 11 for a complete list of mutations. *Frequency includes 2 ARID1A variants rescued after ESP filtering.
Vacuolar ATPase gene mutations are recurrent in FL
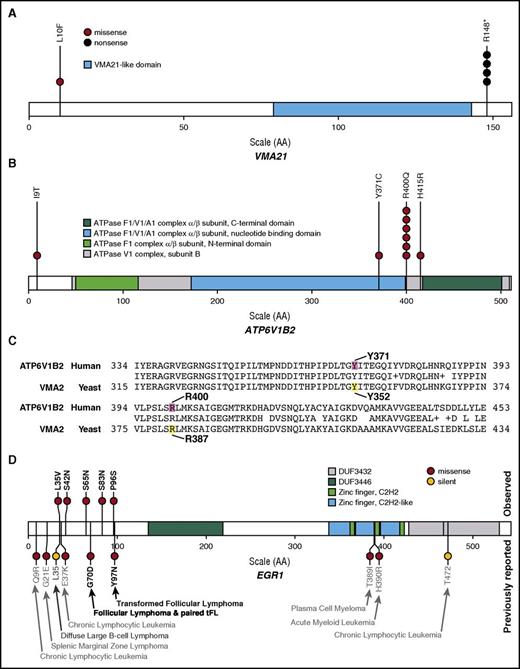
Hotspot mutations were identified in 2 SMGs associated with vacuolar H+ ATPase (V-ATPase) assembly and function: previously described ATP6V1B2 (R400Q)17 and previously unreported VMA21 (R148*; Figure 5A-B). The multi-subunit V-ATPase complexes are responsible for proton transport across membranes, maintain pH homeostasis in organelles, and are localized at the plasma membrane of some cells.57 Mutations in ATP6V1B2 and ATP6AP1 have been described in FL and, along with RRAGC mutations, are hypothesized to alter lysosomal amino acid sensing and downstream mTORC1 signaling.14,17 We observed a similar spectrum of RRAGC mutations to those previously described, although at a lower frequency (5.7%) in our cohort.17 Five patients harbored VMA21 mutations (4 with R148*). VMA21 is involved in V-ATPase assembly and, when lost, results in rapid degradation of the V0 domain.58,59 Nine patients harbored missense mutations in the B2 subunit of the V1 domain protein, ATP6V1B2 (6 with R400Q). Site-directed mutagenesis of the yeast homolog Vma2p at residues Y352S and R381S resulted in complete loss of ATPase activity and proton transport.60 When aligned to human ATP6V1B2, these sites are homologous to 2 locations mutated in our cohort: Y371 and the hotspot R400 mutation (Figure 5C). Finally, we identified additional novel missense mutations in the main V-ATPase complex (ATP6V0A1 [0.95%]; ATP6V1F [0.95%]) as well as an accessory protein (ATP6AP2 [1.9%]). Together, these mutations implicate the loss of, or altered V-ATPase function, in FL pathogenesis.
Recurrent mutations in vacuolar ATPase genes and EGR1 in patients with FL. Hotspot mutations were identified in significantly mutated vacuolar ATPase-associated genes: (A) VMA21 (R148* in 4 of 5 mutated patients; ENST00000370361) and (B) ATP6V1B2 (R400Q in 6 of 9 mutated patients; ENST00000276390). (C) BLAST alignment results illustrating highly conserved yeast Vma2p (YBR127C) amino acid residues previously shown to abrogate ATPase catalytic activity when mutated (yellow) are orthologous to amino acid residues altered by mutations in human ATP6V1B2 (ENST00000276390.2) observed in our cohort (magenta). (D) EGR1 mutations observed in this cohort (N = 105), indicated above the protein diagram, were only observed near the N-terminus of the protein (ENST00000239938). EGR1 mutations previously reported for hematopoietic malignancies in COSMIC and selected papers are depicted below the protein diagram.8,23,24,68,69 See supplemental Table 11 for a complete list of V-ATPase complex and EGR1 mutations.
Recurrent mutations in vacuolar ATPase genes and EGR1 in patients with FL. Hotspot mutations were identified in significantly mutated vacuolar ATPase-associated genes: (A) VMA21 (R148* in 4 of 5 mutated patients; ENST00000370361) and (B) ATP6V1B2 (R400Q in 6 of 9 mutated patients; ENST00000276390). (C) BLAST alignment results illustrating highly conserved yeast Vma2p (YBR127C) amino acid residues previously shown to abrogate ATPase catalytic activity when mutated (yellow) are orthologous to amino acid residues altered by mutations in human ATP6V1B2 (ENST00000276390.2) observed in our cohort (magenta). (D) EGR1 mutations observed in this cohort (N = 105), indicated above the protein diagram, were only observed near the N-terminus of the protein (ENST00000239938). EGR1 mutations previously reported for hematopoietic malignancies in COSMIC and selected papers are depicted below the protein diagram.8,23,24,68,69 See supplemental Table 11 for a complete list of V-ATPase complex and EGR1 mutations.
Hotspot mutations in transcription factor POU2F2 and coactivator POU2AF1
We also identified POU2F2 (OCT2) as significantly mutated with a frequency of 5.7% (6/105), similar to the previously reported 8% frequency15 and containing the same hotspot mutation. The T239 hotspot occurs within helix 3 of the POU-specific domain of this transcription factor, which, in accordance with crystal structure data, is a region critical to binding of the POU-specific binding site.61 In addition, we identified the POU2F2 binding partner POU2AF1 (BOB1/OCAB) as significantly mutated in this cohort (6.7%). POU2AF1 is a B-cell-specific coactivator that works in concert with POU2F2 to promote immunoglobulin transcription and is essential for germinal center formation in mice.62 POU2AF1 also contained a hotspot with 6 of 7 mutations occurring in the 2 bp exon 1 splice donor site (supplemental Figure 11). Although mutations in either POU2F2 or POU2AF1 are relatively rare, the single non-hotspot missense mutation in POU2AF1 (P27A) occurred in a patient who also had a POU2F2 (T239A) mutation.
SWI/SNF complex genes are recurrently mutated in FL
The SWitch/sucrose nonfermentable (SWI/SNF) nucleosome-remodeling complexes modulate chromatin structure and are mutated in an estimated 20% of cancers.63 Although the only SMG in this complex was BCL7A, recurrent mutations in 9 SWI/SNF complex genes were observed, affecting 32 (30.5%) patients in our cohort at mutation frequencies ranging from 0.95% to 19.4% (Figure 4B; supplemental Table 11). Among these was ARID1A (5.7%), a modifier gene in the m7-FLIPI associated with longer failure-free survival.18 These findings indicate that the SWI/SNF nucleosome-remodeling complex is highly and recurrently mutated in FL.
Recurrent EGR1/2 mutations in FL
EGR1 was recurrently mutated in our FL cohort (4.8%), but was modestly above the FDR cutoff (q = 0.0799) used to define our single-gene SMG list. However, these mutations are of interest because this transcription factor cross-regulates and interacts with the CREBBP/EP300 complex, is induced by BCR signaling, and is essential to marginal B-cell development.64-66 In addition, Egr1+/− and Egr1−/− mice are more susceptible to developing T-lymphomas than their wild-type littermates after ENU mutagenesis.67 Analysis of mutations in COSMIC and other recent NHL sequencing studies identified reports of similar N-terminal mutations in patients with splenic marginal zone lymphoma, CLL, diffuse large B-cell lymphoma, and both primary and transformed FL (Figure 5D).8,23,24,68,69 EGR1 was also recently reported as mutated in 2/10 Hodgkin lymphoma samples exome sequenced.47 Another family member, EGR2, which has been implicated in dysregulated BCR signaling in CLL,70,71 was also recurrently mutated in our cohort (1.9%).
Recurrent NOTCH mutations
Recurrent mutations in NOTCH1 and NOTCH2 have been previously described in FL,72 and have been shown to contribute to the etiology of other NHLs.19,22,24,68,73 We observed NOTCH1 (3.8%) and NOTCH2 (3.8%) mutations at a rate similar to that previously described,72 with novel mutations in NOTCH3 (4.8%) and NOTCH4 (4.2%, only evaluable in the discovery cohort), as well as the NOTCH signaling regulators DTX1 (5.7%) and SPEN (2.9%), affecting 19 patients (18.1%).
Clinical outcomes
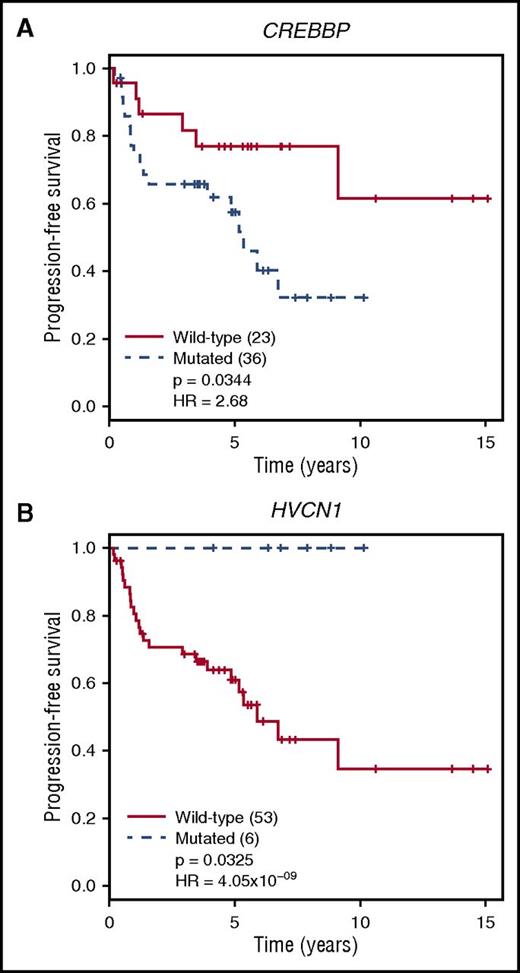
Complete treatment history and outcome data were available for 100 patients. We evaluated the clinical effect of SMGs in a subset of treatment-naive FL samples (N = 82), excluding transformed and relapsed FL samples (Table 1). This cohort reflected the significantly shorter overall survival associated with a high-risk FLIPI score compared with patients with a low-/intermediate-risk score (P = .00275; q = 0.019 after Benjamini-Hochberg correction; supplemental Figure 12A). This difference was also true for the subset of patients that received treatment (excluding observation) within 1 year of diagnosis and the sequenced biopsy (N = 59; P = .00227; q = 0.019) (supplemental Figure 12B). All 7 genes incorporated into the m7-FLIPI score were targeted by our capture design, and were therefore evaluable for our patients.18 Progression-free survival (PFS) was not different for patients classified as high- vs low-risk by m7-FLIPI score in the overall or treatment-only cohorts (supplemental Figure 12C-D); however, the number of patients characterized as high-risk is small (N = 9 or 10). We next evaluated PFS in treatment-only patients stratified by the presence or absence of mutations in SMGs. HVCN1 mutations (N = 6) were associated with improved PFS in treated patients (P = .033; q = 0.740), whereas CREBBP mutations (N = 36) were associated with reduced PFS (P = .034; q = 0.884; Figure 6). When these analyses are extended to include observed patients, HVCN1 mutations (N = 7) show the same trend (P = .058; q = 0.975), and CREBBP mutations (N = 43) remain significantly associated with reduced PFS (P = .029; q = 0.894) (supplemental Figure 13), despite fewer-than-expected CREBBP mutations in the observation group compared to treated groups (supplemental Table 7). These associations are of clinical interest, but will require validation, particularly the more rare HVCN1 mutations, in larger cohorts.
Mutations affecting PFS in treated patients with FL. Treatment-naive patients who received treatment within 1 year of diagnosis and sample collection (N = 59) were stratified by the presence or absence of coding or splice site mutations in SMGs, with a minimum of 5 mutations in this subset of patients (supplemental Table 6). Only groups showing significantly different survival are shown. (A) PFS was worse for patients harboring CREBBP mutations (P = .034; q = 0.884 after Benjamini-Hochberg correction for multiple hypothesis testing). (B) In contrast, patients with HVCN1 mutations had better PFS than those with wild-type HVCN1 (P = .033; q = 0.740).
Mutations affecting PFS in treated patients with FL. Treatment-naive patients who received treatment within 1 year of diagnosis and sample collection (N = 59) were stratified by the presence or absence of coding or splice site mutations in SMGs, with a minimum of 5 mutations in this subset of patients (supplemental Table 6). Only groups showing significantly different survival are shown. (A) PFS was worse for patients harboring CREBBP mutations (P = .034; q = 0.884 after Benjamini-Hochberg correction for multiple hypothesis testing). (B) In contrast, patients with HVCN1 mutations had better PFS than those with wild-type HVCN1 (P = .033; q = 0.740).
Discussion
Recent publications have provided a strong foundation for the identification of important players in the pathogenesis of FL. The current study markedly increases the number of genes known to be mutated in several previously described pathways and protein complexes, as well as identifies a number of novel genes such as BTK, HVCN1, POU2AF1, and ZNF608 for future studies. We describe a much higher overall number and rate of histone gene mutations than previously observed, as well as report the frequent and significant co-occurrence of these mutations within individual patients. In contrast to most prior studies, our cohort consisted primarily of asymptomatic, low-risk, treatment-naive patients with FL, which may be useful in future comparisons with symptomatic and high-risk FL, relapsed or transformed cohorts.
Our approach using other B-cell malignancy studies in combination with a discovery data set to guide our capture design permitted the identification of low-frequency recurrent mutations. This strategy mitigates some biases introduced by using only recurrent mutations identified within a small discovery set for extension in a larger number of samples. Ideally, samples could be flow sorted before sequencing; however, this is not possible for most archival samples. As demonstrated by supplemental Figure 1, tumor purity can reduce the number of detectable mutations, particularly when subclonal, in a given sample. Overall variant recurrence estimations, particularly for mutations not primarily identified in the founding clone, may be underestimated for this reason. To increase purity, FFPE blocks were evaluated by a pathologist, and regions enriched with tumor cells were chosen for sequencing. This method performed well when compared with other data sets. Pastore et al18 evaluated 74 genes in patients with bulky, symptomatic, or advanced-stage disease who received rituximab-based treatment and identified 22 SMGs. Despite differences in patient populations, we also identified 11 of these genes as significantly mutated, with recurrent mutations in all 20 of the 74 genes that were targeted by our custom capture panel. Only 17/39 (43.6%) of our SMGs were evaluated in their study (supplemental Figure 14). Notably, the publically available International Cancer Gene Consortium malignant lymphoma data set identified coding mutations in 31 of our 39 SMGs.
We attempted to evaluate the m7-FLIPI18 for patient stratification in our cohort; however, differences in patient populations resulted in only a few patients ultimately characterized as high-risk. The number of patients with high-risk FLIPI scores was 50% to 51% in the previous study compared with 22% in ours (Table 1), which resulted in only 10 (12.2%) patients overall, and 9 (15.3%) who received treatment who were categorized as high-risk by m7-FLIPI in our cohort. It is not clear whether these results were not significant because of low power or differences in treatment, but they indicate that larger cohorts will be required to validate the m7-FLIPI and determine whether application of this risk stratification is appropriate for additional, particularly lower stage or more heterogeneously treated, patient cohorts.
In addition to expanding the list of genes mutated in known pathways implicated in FL etiology, we identified a number of novel candidate genes such as HVCN1, ZNF608, LRRN3, CCDC129, EGR1, POU2AF1, and BTK. HVCN1, a hydrogen voltage-gated proton channel, acts downstream of the BCR, is a BCL6 target, and is downregulated in proliferating B cells.74 Increased expression of a short isoform of the protein that enhances BCR signaling was shown in patients with CLL compared to healthy donor peripheral blood B cells.75 Mutations affecting proton transporters in both HVCN1 and the V-ATPase proteins affect 21.9% of our cohort and provide additional avenues for investigation into the role of cellular pH in FL pathogenesis. Furthermore, there is likely an underestimate of the frequency of V-ATPase complex mutations, as only ATP6V1B2, ATP6V1F, and ATP6AP2 were targeted by our custom capture reagent, warranting future studies of this complex in FL. Although mechanistic evidence exists supporting roles for EGR1, POU2AF1, and BTK in lymphomagenesis, little is known about ZNF608. Further investigation is warranted, as ZNF608 mutations have been reported in single cases of other lymphomas, including splenic marginal zone, primary central nervous system, and T-cell lymphoma, in addition to a few patients with diffuse large B-cell lymphoma, indicating a more universal role in lymphomagenesis not restricted to FL.76-79
Comprehensive sequencing of iNHLs remains limited, with CLL the best characterized to date. Landau et al69 identified 19 SMGs in CLL, 2 of which overlapped with our SMG list (HIST1H1E, TP53), although we observed mutations in 13/19 of these genes. Larger numbers of patients will be required to appreciate the full complement of overlapping and distinct mutations with functional relevance in these and other iNHLs. These similarities and distinctions may be important for further development of targeted therapeutic agents, as well as the identification of appropriate patient populations for study. Our identification of mutations in BTK, SYK, and proteins that act downstream of these kinases may have implications for the use of emerging targeted therapies such as ibrutinib and entospletinib. In BTK, we identified 8 mutations, 2 of which have evidence to suggest they may confer ibrutinib resistance. In addition, CARD11 coiled–coil domain, BCL10 nonsense, and TNFAIP3 loss-of-function mutations are associated with NF-kappaB activation and, similar to PLCG2 mutations, may render targeting of upstream BTK and SYK ineffective in these patients. Whether these mutations will affect ibrutinib therapy response has yet to be determined, but should be investigated further, as they affected 23 patients (22%) in our cohort. This figure does not include several BCR pathway genes with mutations of unknown significance (Figure 4A) or alterations not readily detected by our methods. Together, it seems likely that a significant number of treatment-naive patients with FL contain ibrutinib-resistant clones before therapy. We were underpowered to evaluate the effects of BTK or SYK mutations on PFS in the context of conventional therapies. We did, however, observe reduced PFS in patients harboring CREBBP mutations, consistent with the enrichment of these mutations in patients identified as having high-risk m7-FLIPI scores.18 Because of the limited number of mutations in TP53 or ARID1A observed in patients evaluated for PFS, we were unable to verify previous reports of reduced and increased PFS, respectively, in these patients.18 We also identified both novel mutations in HVCN1 and an apparent association between these variants and increased PFS. These prognostic findings are interesting but limited by the low number of HVCN1 mutations (N = 6) identified in our treatment-only cohort, and will need to be validated in larger cohorts. No genes were significantly associated with disease progression by 24 months (PFS24), a surrogate of overall survival, although CREBBP and HVCN1 trended as expected (supplemental Figure 15). That only 82 treatment-naive patients, 59 of whom received treatment, were evaluated in this study is a major limitation to these survival analyses. Building on this discovery work, we are expanding the size of our cohort for more complete analysis of clinical correlations and various mutations in future studies.
The clinical heterogeneity of FL remains a challenge to both our understanding of disease pathobiology and the clinical care of patients with FL. Using a hybrid custom capture lymphoma panel derived from both discovery sequencing and published reports in other B-cell malignancies, this study identified novel recurrent mutations in patients with FL, expanded the list of mutated genes in known FL pathways, and identified new pathways associated with FL. Such advancements are key to understanding the pathogenesis of FL and making improvements in therapeutic approaches used in the treatment of patients with FL.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the Siteman Flow Cytometry Core for flow cytometry services, which are supported in part by a National Institutes of Health, National Cancer Institute Cancer Center Support Grant (P30 CA91842). M.G. was supported by a National Institutes of Health, National Human Genome Research Institute grant (K99 HG007940). O.L.G. was supported by a National Institutes of Health, National Cancer Institute grant (K22 CA188163). This work was supported by the Lymphoma Research Foundation Follicular Lymphoma Pathway Award (T.A.F.), the Larry and Winnie Chiang Lymphoma Fellowship (T.A.F., F.G., M.M.B.-E.), and the Siteman Cancer Center Research Development Award (T.A.F.), as well as the Foundation for Barnes-Jewish Hospital (N.L.B.).
Authorship
Contribution: N.L.B., M.G., O.L.G. and T.A.F. conceptualized the study. B.S.W., C.A.M., R.S.F., M.G., O.L.G., and T.A.F. developed the experimental design. F.K., A.F.C., K.R.C., M.M.B.-E., N.L.B. and T.A.F. acquired samples and clinical data. C.C.F. led sequence library construction for data acquisition. K.K., F.G., B.S.W., M.M., C.A.M., L.T., M.G., and O.L.G. performed data analysis. K.K. and F.G. prepared figures and tables. K.K., F.G., and T.A.F. wrote the manuscript with input from M.G., O.L.G., and N.L.B. All authors approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Obi L. Griffith, McDonnell Genome Institute, Campus Box 8501, 4444 Forest Park Ave, St. Louis, MO 63108; e-mail: obigriffith@wustl.edu; Malachi Griffith, McDonnell Genome Institute, Campus Box 8501, 4444 Forest Park Ave, St. Louis, MO 63108; e-mail: mgriffit@wustl.edu; and Todd A. Fehniger, Washington University School of Medicine, Campus Box 8007, 660 S. Euclid Ave, St. Louis, MO 63110; e-mail: tfehnige@wustl.edu.